How to Interpret Patterns of Genetic Variation? Admixture, Divergence, Inbreeding, Cousin Marriage
Two different but important population genetics papers have come out. One is Steven Bray et al. (2010) “Signatures of Founder Effects, Admixture, and Selection in the Ashkenazi Jewish Population.” The other one is Isabel Alves et al. (2012) “Genomic Data Reveals a Complex Making of Humans.”
Dienekes (here) and Razib (here, here and here) offer lengthy discussions of the ways geneticists can tease out effects of admixture, founder effect and selection on human populations writ large and on the history of some “non-trivial” populations such as Ashkenazi Jews.
COMMON DESCENT AND ADMIXTURE
Dienekes takes issue with conventional tree models in which populations neatly branch off from each other in a linear fashion. He argues that tree models and historical reconstructions that derive from them (such as the theory of migration of modern humans out of Africa based on the observation that Africans are most divergent among human populations and that non-African variation is a subset of African variation) “wholly depend on ignoring admixture.” This is imprecise as, in reality, tree models create phylogenetic relationships where they historically did not exist. This can be seen from a side-by-side comparison of a phylogenetic tree from the Ashkenazi Jewish study by Bray et al. (2012, Fig. S2, Suppl. Mat.) and a tree from the American Indian study by Reich et al (2012, Fig. S3, Suppl.Mat.).
On the Bray et al. tree, Ashkenazi Jews (AJ) are an outgroup to Russian, Orcadian, French, Basque, Italian, Sardinian and Tuscan. This creates an impression that AJ are an older, more divergent population that shares common descent with the other members of its clade. A direct historical interpretation of this tree would make AJ the oldest population in Europe. However, this would be a patently false interpretation, and we know it from historical sources. The Reich et al.’s tree on the right is usually interpreted as the depiction of an “obvious” fact that American Indians came from East Asia (anchored in Han), while East Asians came from Africa (anchored in Yoruba). But this tree cannot be interpreted in such a way because we know from the Bray et al. tree that geneticists’ “trees” don’t necessarily depict shared descent between populations, but oftentimes a complex pattern of gene flow (admixture) that may break up an actual phylogeny and re-assemble it into a spurious one.
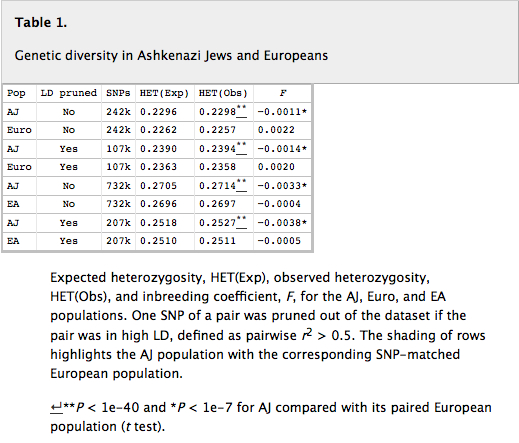
The genetic reality behind the Bray tree is that AJ turned up being more diverse than Europeans (see Table 1 below).
“Surprisingly, we found a higher level of heterozygosity among AJ individuals compared with Europeans (P < 1e-40), confirming speculation made in one recent report and a trend seen in another. Although this difference may appear small, it is highly statistically significant because of the large number of individuals and markers analyzed, even after pruning SNPs that are in high LD. The higher diversity in the AJ population was paralleled by a lower inbreeding coefficient, F, indicating the AJ population is more outbred than Europeans, not inbred, as has long been assumed (P < 1e-7). The greater genetic variation among the AJ population was further confirmed using a pairwise identity-by-state (IBS) permutation test, which showed that average pairs of AJ individuals have significantly less genome-wide IBS sharing than pairs of EA or Euro individuals (empirical P value < 0.05). Thus, our results show that the AJ population is more genetically diverse than Europeans.”
They also found that Linkage Disequilibrium (LD) tends to be higher in AJ than in Europeans. Geneticists often use high LD values in American Indian populations and low LD values in African populations as evidence for a recent Beringian bottleneck in the New World and for Mid-Pleistocene continuity in Africa. But, as Bray et al.’s research shows, since AJ are both diverse and have high LD, LD can reflect not only population antiquity but also a history of mixing between genetically distinct populations. This transpired in other studies as well (see here and here). Hence, the genetic foundation for out-of-Africa and into-the-Americas is shaky and is based on just one way of reading the data.
Bray et al. (2012) also observe that AJ are more diverse than their Middle Eastern relatives:
“We removed SNPs in high LD and measured the mean heterozygosity per locus across the combined Middle Eastern populations (Bedouin, Palestinian, and Druze) and found that the AJ population had higher heterozygosity (0.3121 vs. 0.3053, P < 1e-23).”
This is again surprising because AJ are supposed to be a subset, not a superset, of their geographical source population. Bray et al. resolve the double conundrum by postulating that an originally diverse founding Middle Eastern population admixed with the European population. As a result of the combination of these two factors, AJ ended up being more diverse than their source as well as host populations.
DIVERSITY AS AN INDICATOR OF POPULATION AGE
A heterogeneous source population is an interesting hypothesis. Other studies (e.g., of Idaho Basques and of Bahama Blacks) have demonstrated that migrant populations may end up mimicking very closely the allelic and haplotypic diversity of its source population and even exceeding it. It means that in post-1492 reality we often observe a reversal of the abstract models that geneticists tend to apply to the pre-1492 human population history. A daughter population doesn’t necessarily carry a subset of the variation of its parent population. Bray et al. now suggest that this may be the case for AJ as well. And some geneticists (see Neel, James. “Estrutura populacional de amerindios e algumas interpretacoes sobre evolucao humana,” Origens, Adaptacoes e Diversidade Biologica do Homem Nativo da Amazonia. 1991) warned out-of-Africa theorists from the very beginning that their assumptions of a panmictic ancestral population and a linear decrease in diversity from source to destination are not grounded in actual human population reality. Basically, human migrants are capable of sampling all of the diversity contained in their source population, hence the observed differences in global patterns of variation have everything to do with what happened demographically and selectively to the migrant and the source population after their separation, not with the static degree of common descent between them.
Dienekes is aware of this problem with genetics studies and he takes it almost to my out-of-America extreme of interpretation when he writes,
“Admixture may not only lead us to overestimate divergence between populations: it might lead us to wrongly estimate the directionality of migration itself. Consider a future geneticist, working thousands of years after a collapse of civilization in the near future which led into a breakdown of long-distance travel. Such a scientist would perhaps conclude that the highest genetic diversity is to be found in North America, and conclude that North America colonized the rest of the world.”
Dienekes can be a good student. back in 2009, I wrote on his blog:
“However, taken at face value, America, since 1492, has accrued the greatest levels of allele diversity in the world.”
Science tends to doctor history to focus only on those pieces of it that support the myth it’s trying to prove. Geneticists began modeling human evolution down to Adam and Eve not from 2011, but from 1492 and, by doing so, overlooked the logic of human genetic diversity accumulation that most recent processes have laid bare before their eyes. African American populations, routinely used at the dawn of out-of-Africa theorizing as proxies for native Africans, have been effectively “Native Americans” for several hundred years and, as such, they have made New World (Amerindian plus African American) variation a superset of African variation. A geneticist of the future unfamiliar with the process of European and African colonization of the New World post-1492 would conclude that humans originated in the Americas. And he would be right but for a wrong reason.
ADMIXTURE AND AN AFRICAN ORIGIN OF MODERN HUMANS
Dienekes takes the example of Ethiopia where the mixing of Caucasians and Africans has created an excess in diversity.
“An example of this is Ethiopia. Many studies have presumed to identify a signal of Out-of-East Africa based on diminishing distance from East Africa. But it is completely unclear how this model fares when one takes into account that East Africans are a recently admixed population: their great genetic diversity may be due to the recent intermingling of two very divergent groups of people (Caucasoids and aboriginal East Africans).”
This may be true even for indigenous East Africans (Afroasiatic and Nilo-Saharan speakers), as Poloni et al. (2009) write,
“The strong genetic structure found over East Africa was neither associated with geography nor with language, a result confirmed by the analysis of 6711 HVS-I sequences of 136 populations mainly from Africa. Processes of migration, language shift and group absorption are documented by linguists and ethnographers for the Nyangatom and Daasanach, thus pointing to the probably transient and plastic nature of these ethnic groups. These processes, associated with periods of isolation, could explain the high diversity and strong genetic structure found in East Africa.”
Dienekes’s belief that out-of-Africa is right (“an origin of anatomically modern humans in Africa still seems to be correct”) transcends the actual data. Poloni et al.’s (2009) conclusion is different:
“The high diversity in East Africa was interpreted as a sign of an ancient origin. However, our results might indicate that this high diversity could also come from a particular history of recent migrations and admixture promoted by the pastoralist societies that dominate in the region.”
This fully supports my contention that genetic data is inherently ambiguous as to where modern humans originated. We need to include additional variables into our modeling of genetic processes in order to reconstruct human prehistory beyond ambiguity.
ADMIXTURE, ENDOGAMY, BOTTLENECK AND COUSIN MARRIAGE
Let’s go back to Bray et al.’s (2010) study of AJ and ask a question: How sufficient is the European admixture hypothesis in explaining the higher diversity values among AJ compared to neighboring Europeans? If we assume that Jews and Christians mixed throughout Jewish history, we should also assume, as a null hypothesis, that gene flow was reciprocal. Consequently, admixture levels should be comparable. Bray et al. don’t test this hypothesis, but their admixture explanation only works if Jews have outbred into Europeans much more effectively than Europeans have outbred into Jews. For this they don’t offer any evidence. But if overall their explanation is right, the question arises what kind of Jewish marriage arrangement favored greater intake of European genes? Could it be that Jews have been practicing sexual selection on their European neighbors (or selective ethnic exogamy) mating with only those Europeans who exhibited physical and behavioral traits conducive to increasing Jewish fitness as a group?
If Jews sampled most of genetic diversity from their source population, then the question arises as to the types of marriage rules and post-marital residence practiced by native Middle Easterners, AJ and Europeans. It seems clear that, while non-Jewish native Middle Easterners sampled by Bray et al. (2012) practice patrilateral ortho-cousin (FBD) marriage, which tends to increase homozygosity, AJ are now known to have prescriptive cousin marriage. It’s possible that it’s the relaxation of marriage prescription in the ancestors of AJ that drove their heterozygosity up, while the maintenance of the FBD rule in native middle Easterners have been keeping their heterozygosity values in check. AJ distinctiveness, therefore, comes from their ethnic endogamy (ethnic inbreeding), which allowed them to “mine” their ancestral gene pool in the context of relative reproductive isolation from European neighbors, and not from clan endogamy (clan inbreeding). Consequently, their higher diversity compared to Middle Easterners stems from the latter’s marriage practices, not necessarily from the former’s admixture with Europeans.
Razib accepts the distinction between ethnic and clan endogamy when he writes,
“The Ashkenazi Jewish pattern of lots of short tracts is why services like 23andMe yield so many “relative” matches for individuals of that background. These people share a lot of the same ancestors, but these are often rather far back in time. So, when considering “inbred” as in the products of frequency cousin marriages, Ashkenazi Jews are not inbred in that manner. They don’t exhibit an abnormal level of very long IBD tracts, which is what you get floating around in the population with there are lots of extremely recent common ancestors between one’s two parents. “
Later, he assumes that Amerindians do not practice clan endogamy and that they are inbred because of a founding effect.
“[T]here is inbreeding, and there is inbreeding. People whose parents are siblings or first cousins are genuinely inbred in a way that Amerindians, who went through a bottleneck, are not.”
Razib has a false perception of American Indian genetics. In reality (see Kirin et al. (2010) “Genomic Runs of Homozygosity Record Population History and Consanguinity,” PLoS ONE 5 (11)),
“The Native American groups (sampled from central and south America) stand apart from all others in having the longest genomic stretches of homozygosity for all ROH length categories. The most extreme individual, a Karitiana from the Brazilian Amazon, has a total of ~861 Mb in runs of homozygosity above 500 kb in length, equivalent to one third of the genome.”
This means that American Indian distinctiveness among other human groups comes not from a founding bottleneck on the way to the New World but from long-term kindred endogamy better known as prescriptive cross-cousin marriage. Karitiana, just like the majority of other American Indian tribes from Lowland South America, practice bilateral cross-cousin marriage when a man’s mother’s brother daughter is the same person as his father’s sister’s daughter because his mother’s brother married his father’s sister. Karitiana don’t even have a word for ‘cross-cousin’ because their cross-cousins are invariably ‘spouses’ (see Landin, Mary. Kinship and Naming Among the Karitiana of Northwestern Brazil. M.A. thesis. University of Texas at Arlington, 1989.)
Long-term kindred endogamy, or the obedience to the rule of cousin marriage over many generations results in the fragmentation of a population into a myriad of self-contained demes. On the level of a genetic locus, clan endogamy quickly results in the fixation of a specific allele through drift yielding to high Fst. However, on the level of a population, comprising the total of all the drifting demes, gene diversity will remain stable. As geneticist Michael Krawczak and social anthropologist Robert H. Barnes (“How Obedience of Marriage Rules May Counteract Genetic Drift,” Journal of Community Genetics 1(1): 23–28, 2010) write,
“For both, autosomal and X-chromosomal genes, our simulations revealed that the gene diversity half life is much longer in populations pursuing (highly idealized) cross-cousin mating than in randomly mating populations. In a population of 100 individuals (50 females, 50 males), for example, the median half life of autosomal gene diversity was 250 generations assuming random mating, compared to 644 generations for cross-cousin matings. If the population size equalled 150, the corresponding figures were 378 and 1,283, respectively. The difference was even more pronounced for X-chromosomal genes. For a population of size N =
= 100, the gene diversity half life under matrilateral cross-cousin mating was approximately six times that under random mating (1,616 generations vs 278 generations), and still four times higher than with patrilateral cross-cousin mating (416 generations). If the population size increased to N
100, the gene diversity half life under matrilateral cross-cousin mating was approximately six times that under random mating (1,616 generations vs 278 generations), and still four times higher than with patrilateral cross-cousin mating (416 generations). If the population size increased to N =
= 150, the corresponding figures were 426 (random), 715 (patrilateral), and 3,440 generations (matrilateral), respectively.”
150, the corresponding figures were 426 (random), 715 (patrilateral), and 3,440 generations (matrilateral), respectively.”
For my purposes here, it means that long-term kindred endogamy produces an illusion of population recency. Genetic markers found in a kindred-endogamic population may be very old. The breakdown of a system of cousin marriages may result in the progressive accumulation of diversity through either mutation or admixture coupled with the faster removal of old markers from the population. It’s therefore possible that “phylogenetic” trees are capable of not only masking admixture events but obfuscating the systematic molecular outcomes of the evolving marriage prescriptions.
The breakdown of an ancestral kinship system based on the rule of bilateral cross-cousin marriage is the essence of all major anthropological theories of the evolution of human kinship systems. Alan Barnard is likely mistaken in seeking an alternative “cognatic” prototype among Pygmies and Khoisans, the two populations presented by geneticists as basal to other African and all non-African populations. He’s mistaken because geneticists are mistaken in not studying the anthropological tradition of kinship evolution research. They base their models on the assumption of a panmictic population, but if anthropological evidence is correct, then human population structures are ultimately derived from an originally highly structured population. If a positive marriage rule translates into many generations of inbreeding and thus places a strong constraint on heterozygosity at loci, while maintaining old markers within the population, then in Africa populations such as Hadza characterized by depressed diversities should be considered as retentions from the earliest, Late Stone Age migrants into Africa (see more in my comments discussion here). Khoisans and Pygmies, on the other hand, are likely products of waves of admixture in South and Central Africa. The breakdown of a highly structured ancestral population entails multiple forms of outbreeding, from marriage to relatives more remote than first cousins but still derived from the same ethnic group to extreme forms of outbreeding and ethnic exogamy. Regardless of the magnitude of outbreeding, every instance of outbreeding is essentially a form of diversity-enhancing and Fst-reducing admixture.




Why do you call the Bray et al. “Signatures of Founder Effects, Admixture, and Selection in the Ashkenazi Jewish Population” a ”2012” when it seems to be a ”2010”?
I corrected it to 2010. Thanks for the catch.
[…] of modern human genetic variation and the inferred phylogenetic trees should be different (see more here)]. It’s understandable that Trautmann & Whiteley’s may have been scared away by the […]
It certainly seems you are missing the obvious about AJ history. AJ population did not originate in Eastern Europe, but appears to have travelled up from Spain (and quite possibly other places) but on the way, they were always militarily weak. This subjected them to rape and rapine throughout their European history, and certainly Eastern European history. Further a peculiarity of Jewish lore, ie Jewishness is accepted if the mother is Jewish, would make it possible to ignore paternity when it is convenient to do so (this despite other Jewish lore which has the category of “momza” miss-translated as bastard, but really meaning, one to whom marriage would bring discredit on a family… ). For the record, I have grad degrees in cultural anthropologist, though I made my living as a programmer.