Howler Monkeys, Neandertals, Pygmies, Khoisans and More: Society for Molecular Biology and Evolution 2012
A s I write, Society for Molecular Biology and Evolution (SMBE) is conducting its annual meetings in Dublin, Ireland. Dienekes has many useful pullouts from the available abstracts. I will make short comments on a few of them, plus bring in those that are not highlighted by Dienekes but fit with the theme of this weblog.
s I write, Society for Molecular Biology and Evolution (SMBE) is conducting its annual meetings in Dublin, Ireland. Dienekes has many useful pullouts from the available abstracts. I will make short comments on a few of them, plus bring in those that are not highlighted by Dienekes but fit with the theme of this weblog.
Color vision variation of trichromacy by L/M hybrid genes in wild populations of New World howler monkeys
Yuka Matsushita, Hiroki Oota, Barbara Welker, Mary Pavelka, Shoji Kawamura
Most New World monkeys have highly polymorphic color vision achieved by allelic variation of the single-locus L/M opsin gene on the X chromosome. Among them, howler monkeys (Alouatta) are an exception having presumably a uniform trichromacy achieved by the L/M opsin gene duplication as in Old World primates (Old World monkeys, apes and humans). A previous in vivo (electroretinogram; ERG) measurement of howlers’ visual sensitivity indicated that the sensitivity was explainable if the L and M opsins of howlers had similar spectral sensitivities with those of Old World primates, respectively, and both were expressed. Therefore, howler monkeys have been supposed to have same uniform trichromatic color vision as Old World primates and are important to study the evolutionary condition for trichromacy in primates. However, in fact nucleotide variation of the L and M opsin genes in wild howler populations has not been studied, and spectral sensitivities of howlers’ L and M opsins have not been directly measured. To verify howlers’ uniform trichromacy non-invasively, we examined fecal DNA samples of howlers (A. palliata and A. pigra) collected in Costa Rica, Nicaragua and Belize. We confirmed that the absorption spectra of L and M opsins are equivalent to those of Old World primates by reconstitution of these photopigments in vitro. Surprisingly, we found various types of L/M hybrid gene in the population samples. These hybrid L/M opsins showed the shift of absorption spectra from normal L and M opsins in vitro. This indicates that color vision of howlers should be polymorphic among at least trichromatic phenotypes, prompting us to reconsider an evolutionary significance of uniform trichromacy in primates.
New World monkeys exhibit considerable variation in color vision. It stems from allelic variation of the L/M opsin gene on the X chromosome. I bring up the issue of trichromatic vision in howler monkeys in conjunction with out-of-America I (modern human origins from New World primates with subsequent expansion of modern humans from the Old World to the New World). Matsushita et al. confirmed that howler monkeys have trichromatic vision, just like humans and Old World primates, by obtaining howler DNA and reconstitution the relevant photopigments in vitro. Although I’m not supportive of Out-of-America I, the case of trichromatic vision in New World monkeys is interesting. In one of my conversations with Stanford University’s Dmitry Petrov, he mentioned the absence of trichromatic vision in New World primates as a serious argument against a Platyrrhine source for modern humans.
A genomewide map of Neandertal ancestry in modern humans
Sriram Sankararaman, Nick Patterson, Swapan Mallick, Svante Paabo, David Reich
Analysis of the genomes of archaic hominins, such as Neandertals and Denisovans, has revealed that these groups have contributed to the genetic variation of modern human populations. Yet, we know little about how these ancient mixtures have shaped the genetic structure of human populations and even less about their impact on human evolution. To answer these questions systematically, we need a map of archaic ancestry i.e., a map that labels whether each region of an individual genome is descended from these archaics. Building such a map is technically challenging because of the antiquity of these gene flow events. We have identified signatures based on patterns of variation at single SNPs as well as haplotypes that are informative of ancient gene flow. We propose a principled method based on the statistical framework of Conditional Random Fields (CRFs) that integrates these patterns leading to highly accurate predictions.
We applied our method to polymorphism data in European and East Asian individuals from the 1000 genomes project, in conjunction with the draft sequence of the Neandertal genome, to obtain the first genomewide map of Neandertal ancestry. Analysis of this map reveals several findings:
1. We identify around 35,000 Neandertal-derived alleles in Europeans and 21,000 in East Asians.
2. The map allows us to identify Neandertal alleles that have been the target of selection since introgression. We identified over 100 regions in which the frequency of Neandertal ancestry is extremely unlikely under a model of neutral evolution. The highest frequency region on chromosome 4 has a frequency of Neandertal ancestry of about 85% in Europe and overlaps CLOCK, a key gene in Circadian function in mammals. The high frequency, Neandertal-derived variant is specific to Europeans; it is not very common in East Asians. This gene has been found in other selection scans in Eurasian populations, but has never before been linked to Neandertal gene flow.
3. Several of the Neandertal-derived alleles identified in 1) above are found in the >6,000 SNPs associated with common diseases listed in the NHGRI catalog. These Neandertal derived variants are found to be risk variants associated with obesity and protective variants against breast cancer.
4. We also investigate the possibility of using this map to reconstruct the genome of the introgressing Neandertal. Using the ancestries in Europe and East Asia, we can reconstruct about 600 Mb which we expect to increase with larger samples and additional populations.
An intriguing paper abstract that disappoints due to the lack of American Indians in the sample. Since American Indians show some striking matches with Neandertals in genetic and phenotypic systems, it’s regrettable that geneticists don’t maintain a sharper focus on testing for possible New World retentions of Neandertal features. The paper also confirms the troubling trend of excluding American Indians from worldwide samples, hence eliminating any opportunities to render a fair test to out-of-Africa vs. out-of-America models of human dispersions.
Inference of demographic history and natural selection in African Pygmy populations from whole-genome sequencing data
Martin Sikora, Etienne Patin, Helio Costa, Katherine Siddle, Brenna M Henn, Jeffrey M Kidd, Ryosuke Kita, Carlos D Bustamante, Lluis Quintana-Murci
The Pygmy populations of Central Africa are some of the last remaining hunter-gatherers among present-day human populations, and can be broadly classified into two geographically separated groups, the Western and Eastern Pygmies. Compared to their neighboring populations of predominantly Bantu origin, Pygmy populations show distinct cultural and physical characteristics, most notably short stature, often referred to as the “Pygmy phenotype”. Given their distinct physical characteristics, the questions of the demographic history and origin of the Pygmy phenotype have attracted much attention. Previous studies have shown an ancient divergence (~60,000 years ago) of the ancestors of modern- day Pygmies from non-Pygmies, and a more recent split of the Eastern and Western Pygmy groups. However, these studies were generally based on a relatively small set of markers, precluding accurate estimations of demographic parameters. Furthermore, despite the considerable interest, to date there is still little known about the genetic basis of the small stature phenotype of Pygmy populations. In order to address these questions, we sequenced the genomes of 47 individuals from three populations: 20 Baka, a Pygmy hunter-gatherer population from the Western subgroup of the African Pygmies; 20 Nzebi, a neighboring non- Pygmy agriculturist population from the Bantu ethnolinguistic group; as well as 7 Mbuti, Eastern Pygmy population, from the Human Genome Diversity Project (HGDP). We performed whole-genome sequencing using Illumina Hi-Seq 2000 to a median sequencing depth of 5.5x per individual. After stringent quality control filters, we call over 17 Million SNPs across the three populations, 32% of them novel (relative to dbSNP 132). Genotype accuracy after imputation was assessed using genotype data from the Illumina OMNI1 SNP array, and error rates were found to be comparable to other low-coverage studies (< 3% for most individuals). Preliminary results show relatively low genetic differentiation between the Baka and the Nzebi (mean FST = 0.026), whereas the Mbuti show higher differentiation to both Baka and Nzebi (mean FST = 0.060 and 0.070, respectively). Furthermore, we find that alleles previously found to be associated with height in other populations are not enriched for the “small” alleles in the Pygmy populations. We find a number of highly differentiated genomic regions as candidate loci for height differentiation, which will be verified using simulations under the best-fit demographic model, inferred from multi-dimensional allele frequency spectra using DaDi. Our dataset will allow a detailed investigation of the demographic history and the genomics of adaptation in these populations.
A clear example of consistency between linguistic classifications and genetic distances. Baka and Njebi (Nzebi) languages coalesce at the level of the Volta-Congo subgroup of Niger-Congo languages. Lese (or the language of the Mbuti Pygmies) is part of the Nilo-Saharan family of languages, which is unrelated to Niger-Congo, in more traditional African classifications, or forms the hypothetical Congo-Saharan macrophylum. Interdisciplinary evidence, as I argued elsewhere, favors common origin between Pygmies and their agricultural neighbors and offers little support for the thesis of supreme antiquity of the Pygmies (linguistically, they are co-eval with farmer languages) or for the hypothesis of language replacement in Pygmies. Alternatively, we may need to consider the possibility that the shallow time depths that linguists allow for the origin of first-order language families (such as Niger-Congo) are hopelessly long, and in reality Niger-Congo is of Mid-Pleistocene age. While the proponents of Paleolithic Continuity theories will certainly welcome such an interpretation, it flies in the face of all of mainstream linguistics as well as archaeology, which has documented the spread of Bantu farmers in the past 5,000 years, which accounts for the vast majority of languages spoken by the Pygmies.
Adaptive evolution of loci covarying with the human African Pygmy Phenotype
Isabel Mendizabal, Urko M. Marigorta, Oscar Lao, David Comas
African Pygmies are hunter-gatherer populations from the equatorial rainforest that present the lowest height averages among humans. The biological basis and the putative adaptive role of the short stature of Pygmy populations has been one of the most intriguing topics for human biologists in the last century, which still remains elusive. Worldwide convergent evolution of the Pygmy size suggests the presence of strong selective pressures on the phenotype. We developed a novel approach to survey the genetic architecture of phenotypes and applied it to study the genomic covariation between allele frequencies and height measurements among Pygmy and non-Pygmy populations. Among the regions that were most associated to the phenotype, we identified a significant excess of genes with pivotal roles in bone homeostasis such as PPPT3B and the height associated SUPT3H-RUNX2. We hypothesize that skeletal remodeling could be a key biological process underlying the Pygmy phenotype. In addition, we showed that these regions have most likely evolved under positive selection. These results constitute the first genetic hint of adaptive evolution in the African Pygmy phenotype, which is consistent with the independent emergence of the Pygmy height in other continents with similar environments.
This is another paper confirming my view that Pygmy phenotype has emerged more than once in human populations. It’s not a primordial human condition retained in Africa and in “pockets” of “living fossils” (such as Andaman islanders) along the putative “southern route” out-of-Africa. Similarly, within Africa, Pygmy populations don’t represent Mid-Pleistocene relics who adopted the languages of their agricultural neighbors, but independent evolutions of Pygmy phenotypes from their respective bases in mid-to-late Holocene Niger-Congo and Nilo-Saharan speech communities.
Population structure and evidence of selection in the Khoe-San and Coloured populations from southern Africa
Carina Schlebusch, Pontus Skoglund, Per Sjödin, Lucie Gattepaille, Sen Li, Flora Jay, Dena Hernandez, Andrew Singleton, Michael Blum, Himla Soodyall, Mattias Jakobsson
The San and Khoe people currently represent remnant groups of a much larger and widely distributed population of hunter-gatherers and pastoralists who had exclusive occupation of southern Africa before the arrival of Bantu-speaking groups in the past 1,200 years and sea-borne immigrants within the last 350 years. Mitochondrial DNA, Y-chromosome and autosomal studies conducted on a few San groups revealed that they harbour some of the most divergent lineages found in living peoples throughout the world. We used autosomal data to characterize patterns of genetic variation among southern African individuals in order to understand human evolutionary history, in particular the demographic history of Africa. To this end, we successfully genotyped ~ 2.3 million genome wide SNP markers in 220 individuals, comprising seven Khoe-San, two Coloured and two Bantu-speaking groups from southern Africa. After quality filtering, the data were combined with publicly available SNP data from other African populations to investigate stratification and demography of African populations. We also applied a newly developed method of estimating population topology and divergence times. Genotypes and inferred haplotypes were used to assess genetic diversity, patterns of haplotype variation and linkage disequilibrium in different populations. We found that six of the seven Khoe-San populations form a common population lineage basal to all other modern human populations. The studied Khoe-San populations are genetically distinct, with diverse histories of gene flow with surrounding populations. A clear geographic structuring among Khoe-San groups was observed, the northern and southern Khoe-San groups were most distinct from each other with the central Khoe-San group being intermediate. The Khwe group contained variation that distinguished it from other Khoe-San groups. Population divergence within the Khoe-San group is approximately 1/3 as ancient as the divergence of the Khoe-San as a whole to other human populations (on the same order as the time of divergence between West Africans and Eurasians). Genetic diversity in some, but not all, Khoe-San populations is among the highest worldwide, but it is influenced by recent admixture. We furthermore find evidence of a Nilo-Saharan ancestral component in certain Khoe-San groups, possibly related to the introduction of pastoralism to southern Africa. We searched for signatures of selection in the different population groups by scanning for differentiated genome-regions between populations and scanning for extended runs of haplotype homozygosity within populations. By means of the selection scans, we found evidence for diverse adaptations in groups with different demographic histories and modes of subsistence.
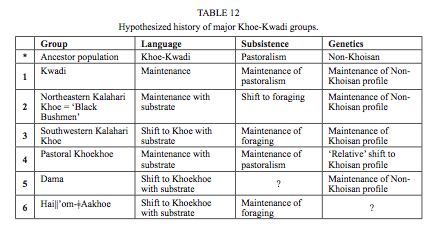
There are three important points here. First, Khoisans apparently remained internally undifferentiated for some 100,000 years (the authors don’t give the divergence estimates but judging by other molecular clock estimates it should be serve a good ballpark figure) after they had split from other humans. This is one of the absurd inferences that come out of population genetic models of modern human origins. Translating it into a more plausible picture, Khoisans began to differentiate among themselves some 40-60,000 years ago, which is to be expected from a population that entered a new continent. Second, the authors caveat the widely publicized notion of world highest intragroup diversity among the Khoisans by pointing out that admixture played a role in boosting Khoisan diversity. Archaic admixture is one possibility. The cumulative effect of multiple migrations into South Africa is another possibility. Third, the authors confirm one such migration, namely the one associated with the spread of pastoralism. Linguistic evidence strongly supports the model whereby the Khoe-Kwadi family entered the Kalahari Basin after the two other families, Ju-ǂHoan and Tuu, and introduced a food-producing economy into the foraging world. Tom Güldemann (“A linguist’s view: Khoe-Kwadi speakers as the earliest food-producers of southern Africa,” Southern African Humanities 20 (2008), 125) described several patterns of maintenance, admixture and acculturation among the Khoe-Kwadi across linguistics, economy and genetics (see below, Table 12):
Population structure and demographic pre-history of the Solomon Islands
Ana Duggan, Mark Stoneking
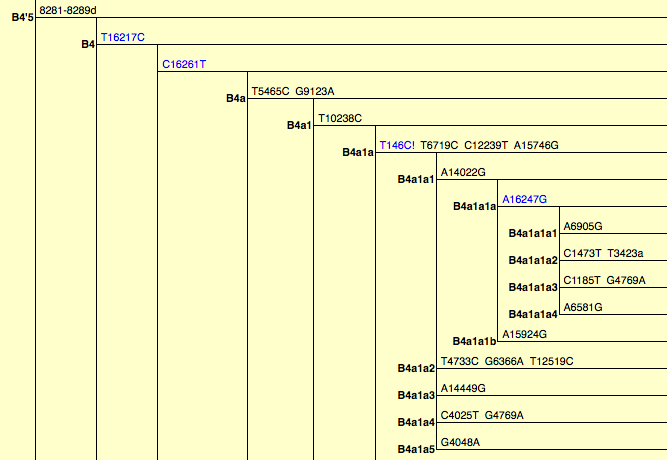
The Solomon Islands are unique in both their geographical situation and population composition. This collection of islands stretch through Near Oceania and into Remote Oceania, the area that spans the boundary of these two regions is both rich in diversity and understudied from a population genetics research perspective. The region is comprised of populations with varied histories, Papuan populations are thought to be descendant from the first peopling of Sahul ~50kya, the Austronesians settled in many coastal areas following their expansion through Island South East Asia ~3kya, and Polynesian Outliers are the most recent settlers, descendants of populations who back-migrated after colonizing Polynesia. The region thus encompasses great variety in population histories, cultures and languages and provides an excellent opportunity to study the effects of multiple migrations and population admixture. Extensive archaeological and linguistic research in the area offers additional sources for comparison and support of genetic findings. Our study has used next-generation sequencing technology to obtain the complete mitochondrial genome of more than 700 individuals from 18 groups spanning Austronesian, Papuan and Polynesian Outlier populations, making it both the largest and most fine grained study of the region to date. Results show that despite retaining language and culture, Papuan populations have mixed substantially with and have no greater Near Oceanic ancestry than Austronesian populations. In striking contrast to all other groups, the Austronesian population from Santa Cruz Islands shows significant Near Oceanic ancestry that confirms archaeological inferences of their unique settlement and early relationship with the Bismarck Archipelago more than 2000km to the Northwest. The mtDNA analyses of Polynesian Outlier groups indicate both recent origins and bottlenecks in these populations in confirmation of their oral traditions. Additionally, we have identified a position in the control region with an unusual mutation pattern; diagnostic for the B4a1a1a haplogroup, the transition at position 16247 appears to have arisen recently and then reverted to the ancestral form several times.
I noted in the past (here and here) that mtDNA phylogenies are built on untested assumptions about ancestral states and homoplasies. Duggan & Stoneking report another case when a unique mutation (A16247G) turns out to be a hot spot showing multiple instances of reversions to the ancestral state. Here is a snapshot of the region of the mtDNA phylogeny containing the B4a1a1a lineage in question (from PhyloTree).
Genetic structure in North African human populations and the gene flow to Southern Europe
Laura R Botigué, Brenna M Henn, Simon Gravel, Jaume Bertranpetit, Carlos D Bustamante, David Comas
Despite being in the African continent and at the shores of the Mediterranean, North African populations might have experienced a different population history compared to their neighbours. However, the extent of their genetic divergence and gene flow from neighbouring populations is poorly understood. In order to establish the genetic structure of North Africans and the gene flow with the Near East, Europe and sub-Saharan Africa, a genomewide SNP genotyping array data (730,000 sites) from several North African and Spanish populations were analysed and compared to a set of African, European and Middle Eastern samples. We identify a complex pattern of autochthonous, European, Near Eastern, and sub-Saharan components in extant North African populations; where the autochthonous component diverged from the European and Near Eastern component more than 12,000 years ago, pointing to a pre-Neolithic ‘‘back-to-Africa’’ gene flow. To estimate the time of migration from sub-Saharan populations into North Africa, we implement a maximum likelihood dating method based on the frequency and length distribution of migrant tracts, which has suggested a migration of western African origin into Morocco ~1,200 years ago and a migration of individuals with Nilotic ancestry into Egypt ~ 750 years ago. We characterize broad patterns of recent gene flow between Europe and Africa, with a gradient of recent African ancestry that is highest in southwestern Europe and decreases in northern latitudes. The elevated shared African ancestry in SW Europe (up to 20% of the individuals’ genomes) can be traced to populations in the North African Maghreb. Our results, based on both allele-frequencies and shared haplotypes, demonstrate that recent migrations from North Africa substantially contribute to the higher genetic diversity in southwestern Europe.