The Western Stemmed Tradition, Clovis and mtDNA 9-bp Deletion
Science 13 July 2012: Vol. 337 no. 6091 pp. 223-228 DOI: 10.1126/science.1218443
Clovis Age Western Stemmed Projectile Points and Human Coprolites at the Paisley Caves
Jenkins, Dennis L. et al.
The Paisley Caves in Oregon record the oldest directly dated human remains (DNA) in the Western Hemisphere. More than 100 high-precision radiocarbon dates show that deposits containing artifacts and coprolites ranging in age from 12,450 to 2295 14C years ago are well stratified. Western Stemmed projectile points were recovered in deposits dated to 11,070 to 11,340 14C years ago, a time contemporaneous with or preceding the Clovis technology. There is no evidence of diagnostic Clovis technology at the site. These two distinct technologies were parallel developments, not the product of a unilinear technological evolution. “Blind testing” analysis of coprolites by an independent laboratory confirms the presence of human DNA in specimens of pre-Clovis age. The colonization of the Americas involved multiple technologically divergent, and possibly genetically divergent, founding groups.
An important paper that continues the business of debunking the Clovis-I model of the the origins of American Indians. Paisley Caves are located in south-central Oregon, hence the name “Western Stemmed Tradition.” The University of Oregon has been conducting excavations at the site since the 1930s but it’s only in 2002 that these 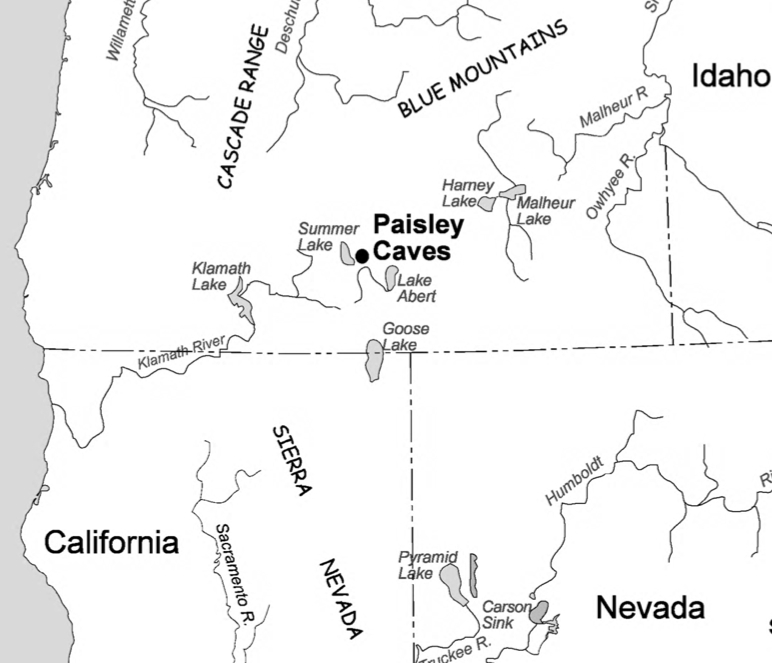 excavations became thorough and consistent. 190 conventional and AMS radiocarbon dates were obtained from coprolites, sedimented urine, artifacts, bones and sagebrush twigs. The assemblage and the human coprolites were dated at 11,070 and 11,340 radiocarbon YBP. confirming that the site is either contemporaneous with or older than Clovis. There were no Clovis-type points found on the site:
excavations became thorough and consistent. 190 conventional and AMS radiocarbon dates were obtained from coprolites, sedimented urine, artifacts, bones and sagebrush twigs. The assemblage and the human coprolites were dated at 11,070 and 11,340 radiocarbon YBP. confirming that the site is either contemporaneous with or older than Clovis. There were no Clovis-type points found on the site:
“There are no classic Clovis technology prismatic blades, overshot flake terminations, overshot flake scars on exceptionally large bifaces, fluting flakes or flute repair flakes in the Paisley assemblage. Rather, the assemblage is dominated by small flake debris reflecting simple core reduction and finish-tool-shaping or rejuvenation by pressure flaking. (Jenkins et al. 2012, Suppl. Mat., 34).
Technologically, the Western stemmed projectile points are different from Clovis points in the following ways (Jenkins et al. 2012, 224):
“Western Stemmed projectile points are generally narrow bifaces with sloping shoulders, and many have relatively thick contracting bases. They were commonly made on flakes by broad collateral, midline, percussion flaking and finished by pressure flaking. In this, they are morphologically and technologically distinct from the generally broader, concave-based, fluted Clovis points made on large bifacial preforms often thinned by overshot flake technology. Prismatic blades—long, narrow flakes with triangular cross sections driven from specially prepared cores—are common to Clovis sites outside of western North America and are less common to WST assemblages.”
Regarding Siberian affinities of the Western Stemmed Tradition, Jenkins et al (2012, 223-224) note,
“Stemmed points were present earlier in East Asia and Siberia, and the basic form could have arrived in the Americas before Clovis developed. Like Clovis, the WST is a New World development sharing basic morphological and technological characteristics with Old World forms.”
Two flake-based projectile point traditions coexisted in Late Pleistocene North America, neither of them have immediate antecedents in Siberia but both of them have generic Old World associations. Importantly, just like with Clovis sites, there is no evidence of microblades at Paisley Caves. Microblades are widely present in Siberia since 20,000 BP and are found in Alaska and along the Pacific Northwest all the way down to the Vancouver Island. But they don’t penetrate any further south suggesting that the areas south of the ice shields were occupied by populations with “New World technological developments,” and not descendants of Siberian migrants. Regarding Clovis, it’s also becoming increasingly apparent that Clovis has a southern origin in North America (the Buttermilk Complex in Texas) and spread north into Alaska and, possibly, Northeast Asia with the retreat of the glaciers. Time will show if the Western Stemmed Tradition, too, expanded up north along the Pacific Coast. For now, Jenkins et al (2012, 227) suggest that the Western Stemmed Tradition was an “indigenous development in the far western United States, whereas Clovis may have developed independently in the Plains and Southeast.
Of special interest is mtDNA obtained from 26 Paisley Caves coprolites. It’s the oldest DNA in the Western Hemisphere (the upper bound is 12,265 ± 25 radiocarbon YBP) and it furnished a surprising finding (see Table S12 in the Supplement):
“Twenty-one specimens belonged to mitochondrial founder haplogroup A, screened for the presence of the A – G SNP at nucleotide position (np) 663 (all positions given with reference to the revised Cambridge Reference Sequence). Five belonged to haplogroup B, characterized by a 9 base-pair (bp) deletion following np 8281, also present in eight of the coprolites assigned to haplogroup A. The presence of this 9bp deletion after nucleotide 8281 has been previously reported for haplogroup A, but it has also been reported for haplogroups C and D.”
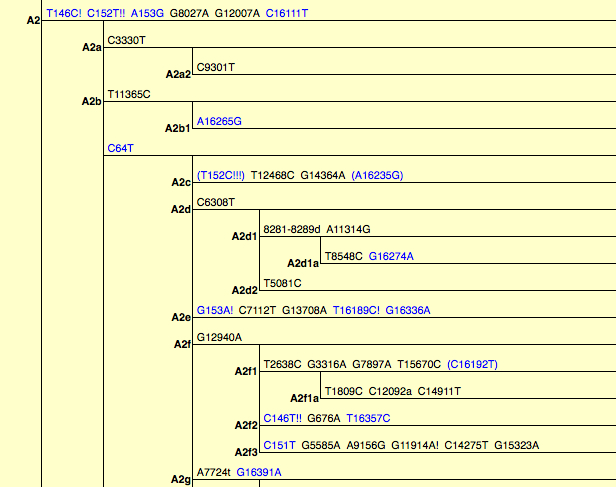
PhyloTree assigns these 9-bp deletion-carrying A lineages to hg A2d1 (see below, partial view) and it’s possible that the Paisley Caves samples are A2d1.
As I reported earlier, 9-bp deletion has a wider distribution globally than just hg B. Geneticists tend to interpret the pervasiveness of this mutation as a sign that it emerged multiple times in human history but this explanation is not particularly compelling. First, methodologically, this may be true in some cases but it doesn’t mean that every time we encounter this deletion on a different haplotypic background, it must be homoplastic. In some cases, it’s the haplogroups that may not have been properly defined. For instance, PhyloTree postulates that the 9-bp deletion emerged independently on hg B6 (supposedly related to hg R11 with no deletion) and B4’5 but it’s just as likely that B6, B4 and B5 form hg B4’B5’B6 to the exclusion of hg R11. Second, the well-established hg B4’B5 is defined as a monophyletic cluster by a combination of 8281-8289d and 16189C but this pair of mutations is found in Africa on L0a2 lineages, in West Asia on T2f lineages and in America on A2d1. In the latter case, A2d1 and B4’B5 are part of macrohaplogroup N. Third, the global presence of 9-bp deletion may indicate great antiquity, and we need to seriously consider the possibility that this mutation dates back to the time prior to the formation of the respective haplogroups. The fact that at Paisley Caves there are more hg A samples with the deletion than hg B samples suggests that A lineages with 8281-8289d may have been rather common in the past.
Distributional evidence supports the antiquity of 8281-8289d or of the 8281-8289d + 16189C combination: it’s rare to non-existent in Siberia but it penetrates deep into South America. If hg B2 was brought to America as a separate, later migration, then we would have found it in Siberia and not likely in South America. The distributional pattern of 9-bp deletion, namely its presence in South America and Sub-Saharan Africa but not in Siberia is not unique. HTVL virus shows the same puzzling geography.


Another point – a ‘C’ at 16189 is the original human form rather than a “mutation”. It is a difference from the CRS but not a difference from the new reference sequence.
With regards to this: “If hg B2 was brought to America as a separate, later migration, then we would have found it in Siberia and not likely in South America.” I do not follow your logic. New World haplogroup B probably came from the region around Japan. It did not *have* to come from Siberia. Some of its highest South American frequencies are found on the western coast i.e. Aymara (83%).
A fair point about 16189C. I paused before typing “mutation” but typed it anyway.
“New World haplogroup B probably came from the region around Japan.”
What do you mean by region around Japan? Japan is surrounded by water.
I can see how hg B can be construed as a circum-Pacific marker. Because of the Andes, because of Polynesia. And I agree that in the Old World it’s definitely an east-of-India marker. But I won’t be assumptive about a “coastal migration into the Americas.” California is not rich in hg B lineages. A coastal migration is a faint possibility that will always be hard to illustrate archaeologically because of the preservation conditions along the coasts.
Hg B is too deep in South America to warrant a separate migration. And it’s too infrequent in East Asia (and non-existent in Northeast Asia) to indicate recency.
Rethinking 8281-8289d + 16189C as a relic combination of states cutting across several haplogroups (but strongly associated with mhg N outside of Africa) and preserved in pockets from Pygmies to Aymara but lost across a wide swath of Eurasia and Australia makes it a broad enough generalization that will help us avoid hasty and murky conclusions about “a coastal migration” carrying hg B2 to America from a region around Japan” for which there’s little to no direct evidence.
By the “region around Japan”, I meant Japan and the adjacent mainland.
The separate haplogroup B migration theory goes with the separate Western Stemmed projectile point technology and the coastal migration theory. The finding of only two types of mtDNA sequences in Paisley caves (A & B) is just icing on the cake.
When the earliest Europeans arrived in the Americans, they considered California to be the most linguistically-diverse portion of North America and so quite a bit of mixing appears to have occurred there. Even with that, though, the Chumash, who have a high frequency of mtDNA haplogroup A sequences ‘stick out like a sore thumb’ among California Indians.
With regards to your statements: “Hg B is too deep in South America to warrant a separate migration. And it’s too infrequent in East Asia (and non-existent in Northeast Asia) to indicate recency.”
Who said anything about “recency”? The earliest dated haplogroup B sequence from Paisley Caves was 12,265 BP (See the supplementary file). Other early haplogroup B sequences have been reported previously as well.
“Japan and the adjacent mainland”
The mainland adjacent to Japan and proximate to America is northeast Asia/eastern Siberia where hg B is rare to non-existent.
“the Chumash, who have a high frequency of mtDNA haplogroup A sequences ‘stick out like a sore thumb’ among California Indians.”
Not sure exactly what you mean with here but this made me think of the following: do hgs A and B tend to co-occur geographically in western New World, as they do in the Paisley Caves? I know hg A is at high frequencies in northwest Canada and Alaska, is it the same further down south, or the Chumash is an exception? With hg X restricted to North America and achieving high frequencies in some Western tribes such as Yakima and Nootka (with eastern distributions likely explained via the eastward Algonquin migration), I wonder if the three Amerindian mhg N clades (X, A and B) tend to cluster the West.
“Who said anything about “recency”? The earliest dated haplogroup B sequence from Paisley Caves was 12,265 BP (See the supplementary file). Other early haplogroup B sequences have been reported previously as well.”
That’s “recent” by my standards. I’m talking about roughly 40,000 for hg B in America.
Adjacent to Japan = to the west (Amur River Region). The only reason I mentioned it is because there have been interactions between the peoples of this region and Japanese.
The Chumash haplogroup A frequency exceeds 50% and they have no ‘B’. The other California Indians, on the other hand, have a haplogroup A frequency of only 6% and 42% ‘B’. Another obvious distinguishing factor between the Chumash and other California Indians is that the former had D4h3 sequences (like OYK Cave man’s) at a frequency of 20% while the others did not. For these reasons (and maybe a few more), the Chumash mtDNA sequences rightly belong with those of the northern populations rather than those of their neighbors.
With regards to the distribution of haplogroups A & B in North America, see Figure 1 (haplogroup A) and Figure 2 (haplogroup B) here:
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC379119/
(Black means low frequency while white/gray means high – this is not overly clear)
Since the above article is not too recent (2002), you can also see many haplogroup frequencies in the supplementary excel file of Raff et al. (2011):
http://onlinelibrary.wiley.com/doi/10.1002/ajpa.21594/suppinfo
Thanks Gisele. I don’t like the maps in the Malhi article, but the paper did answer my question.
For the record researchers have yet to find hg B in ancient human remains from the Old World predating the arrival of Polynesians 3,600-4000 y.b.p. (T. G. Schurr personal communication and every ancient mtDNA reports of Asians including remains of the earliest Lapita Cuture >4,000 ybp. The important work contained in Terrell et al. 1997 are based on multidisciplinary observations that initiate a change in the “given” dynamics of Polynesian history; that their migrations were not chance encounters with little if any subsequent contact following new discoveries of previously unknown atolls. Several points should be highlighted, 1) that there is little if any biological support for a southeast Asian source for the Polynesians, 2) that there was substantial knowledge of and verifiable interisland contact during and following colonization, 3) that the relationship between coastal Melanesians and Polynesians — conformed with an advanced maritime cultural proficiency — suggests that the original Pleistocene inhabitants of Oceania were pushed back into the interior. If the original Pleistocene inhabitants of Oceania were pushed back into the interior — and Polynesians did not have a southeast Asian origin — genetic affinities could link them with South American Indians. Further genetic, linguistic, and cultural links between coastal cultures of Borneo and the original inhabitants of Madagascar could indicate that the explorations of the Polynesians was not limited to the Pacific and may have included the Arabian, Red, and Mediterranean Seas (Phoenicians ?). There are numerous bio-genetic traits shared by South American Indians and coastal Melanesians. Heyerdahl’s book; Native Americans in the Pacific 1952 identifies many archaeological links to the New World that should now transcend, given the overwhelming genetic relationship binding these “first People”, an Amerindian source for the first Polynesians as NOT a migration out of Asia. That the same genetically related people would coincidentally migrate into separate un-peopled areas tens of thousands of years apart is as unfathomable to me as is the likelihood that Polynesians “first peopled” the remote Islands of the Pacific from the Americas, is for others.
Based upon complete sequencing, Native American ‘B’ sequences belong to sub-group B4b and Polynesian to B4a. That, combined with the different Y chromosome haplogroups found in both populations, makes it difficult to trace Polynesians back to the the Americas… Furthermore, all of the ‘B’ branches are only found in the Old World *except* Native American B2 (< B4b).
With regards to early mtDNA haplogroup B sequences in the Old World – in the article entitled, "Intracemetery Genetic Analysis at the Nakazuma Jomon Site in Japan by Mitochondrial DNA Sequencing" Shinoda et al. reported a sequence which can be classified as B4b1a1 today. Yao et al. (2002) wrote the following on it:
"An interesting case is constituted by the 29 mtDNAs from the 4,500-year-old Nakazuma Jomon site that were sequenced for the region 16209–16402 (Shinoda and Kanai 1999). …type 7 (16284) matches a B4b haplotype from Liaoning…."
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC384943/
Take a look at haplotypes T2CH711 & T2CH712, though, in Table 1 of the following article:
Ancient DNA reveals kinship burial patterns of a pre-Columbian Andean community
Baca et al. (2012)
http://www.biomedcentral.com/1471-2156/13/30
The mutations of these two HVR I hapotypes are identical to the Polynesian motif but the authors determined, "all B4a SNPs were CRS and our multiplex indicated that these samples belong to B2d haplo." (personal communication) . I find the B2d classification hard to believe but what can I say?
I have a relevant pub and am very interested in this blog
Faught, Michael K
2008 Archaeological Roots of Human Diversity in the New World: A Compilation of Accurate and Precise Radiocarbon Ages from Earliest Sites. American Antiquity 73(4):670-69.
Also you may find Sassaman 2010 a great read
Sassaman, Kenneth E.
2010 The Eastern Archaic, Historicized. Issues in Eastern Woodlands Archaeology. Altamira Press, Lanham.
mkf
Hi Michael, thank you for the references, welcome to Anthropogenesis. Your paper looks like it is an especially urgent one for me to read – am trying to keep up with all the literature but in vain, so thanks for the pointer – and the Sassaman book is an interesting one, too. I may write a post about your paper once I read it. Stay tuned!
Geman, I note in that 2008 paper a sympathy with the Clovis = Solutreans alternative, but now I understand the phylogenies better, X is Asiatic, not Euroopean (even if it shows up there). Also NB “Solutrean” remains with U and R0 haplogroups at http://www.eupedia.com/europe/ancient_european_dna.shtml
I like the idea of the “still stand” in South America, everthing comes up and out of there throughout the Holocene, earliest domesticates, earliest complex societies, most linguistic diversity, and all five haplogroups. cf Wang et al 2007, Fagundes 2008, Tamm et al 2007, O’Rouke and Raff 2010.
Just sharing
I agree completely that hg X is no evidence for the Solutrean origin of Clovis. And by “Asiatic” you must be meaning “West Asian,” which is a very unusual place to find relatives of Amerindian lineages. The geographic footprint left by hg X is reminiscent of another puzzling pattern observed across different genetic systems, namely a connection between Amerindians and Europeans (mtDNA hg B in America and hg U in Western Europe, both part of mhg R; Y-DNA P clade represented by Q in America, West Asia (again!) and Northern Europe and R in Europe; Amerindian “admixture” in Europe detected in autosomes). In all these cases, East Eurasia, albeit adjacent to the New World geographically, doesn’t show this signal with the same intensity, if at all. Ultimately, the mtDNA hg X phenomenon and the exclusive Amerindian-European connections may very well be relics of the same ancient population process.
I recently came across this genetics study of a living native californian, and several historic remains from the Monterey pennisula.
http://pcas.org/assets/documents/PagesfromV40N1a.pdf
[…] are fed data and arguement like this you must wonder if someone is blowing smoke on purpose. I do The Western Stemmed Tradition, Clovis and mtDNA 9-bp Deletion As I reported earlier, 9-bp deletion has a wider distribution globally than just hg B. Geneticists […]