Ancient Molecular and Morphometric Variation from Chile: The Death of the Dual-Entry Model?
Chungara, Revista de Antropología Chilena Vol. 43, No 2, 2011, pp. 283-292
Morphometric and mtDNA Analyses of Archaic Skeletal Remains from Southwestern South America
Germán Manríquez, Mauricio Moraga, Calogero Santoro, Eugenio Aspillaga, Bernardo T. Arriaza, and Francisco Rothhammer.
For decades anthropologists have discussed how and when the Americas were peopled. The prevailing view is that the first Paleoindians, ancestors of the Amerindians, arrived from Asia and Beringia to the American continent using a Pacific coastal route in pre-Clovis times. In this article skeletal remains dated 9000-4000 BP, excavated from archaeological sites in northern, central and southern Chile, were analyzed using geometric morphometric and ancient mtDNA techniques. Results indicate that the ancient cranial material from southwestern South America exhibit a wide range of cranial vault shape variation which is independent of chronology. mtDNA restriction and sequence analysis performed on the same skeletal remains, revealed only the presence of the main four founding mtDNA haplogroups (A, B, C and D) as early as 9,000 BP. Our results using morphometric and molecular mtDNA haplogroup data show that human populations inhabiting the Americas during archaic times can not be considered as belonging to two different groups on the basis of analyzed data. These results are consistent with those recently obtained using complete sequence mtDNA analyses.
Link (Free Access PDF)
It’s a pleasure to blog about research published in lesser known scientific journals. Data published in peripheral sources is often unknown to the mainstream academic and amateur public in the U.S. and Europe. Introducing Chungara, a Chilean anthropological forum, and an article reporting on morphometric and mtDNA variation in a sample of 27 Early, Middle, and Late Archaic skulls from Northern, Central and Southern Chile dated at 9000-4000 BP. There are two key findings reported in this paper.
1. All four principal American Indian mtDNA markers (hgs A, B, C, D) were detected in the sample, which indicates the stability and diversity of genetic composition of South American Indians in pre-agricultural times. At the same time, haplogroup assignment is rather crude, as sublineage-specific SNPs weren’t screened for. Hg B was reported for the first time south of 43 degrees south latitude. It means it was present in the area by at least 8,000 BP but was subsequently lost there, as modern populations in southern South America don’t have it. This finding doesn’t make life easier for those scholars who continue to derive hg B from a separate migration into the New World, although in North America hg B is attested at 12,000 YBP (Paisley Cave). Also, previously hg A has never been reported in ancient remains earlier than 4,504 YBP.
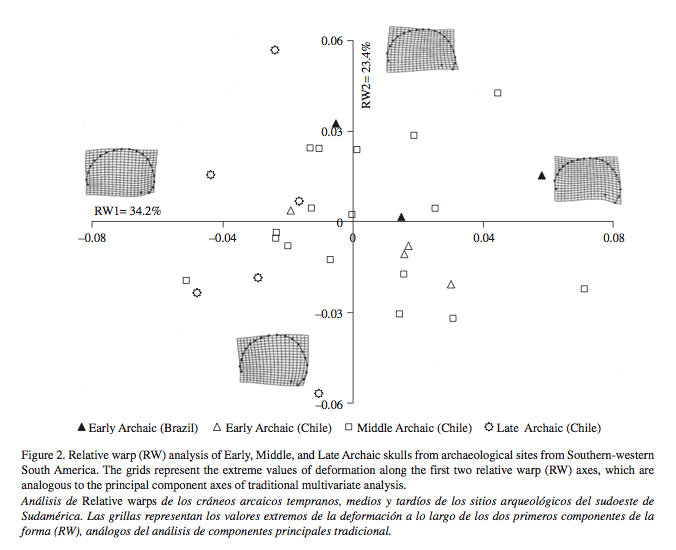
2. Manríquez et al. (2012) failed to detect a division of skulls into two broad groups – the earlier “Australomelanesian” type and the later “Mongoloid” type. This division has been championed in the late 1990s-early 2000s by multiple teams of paleobiologists who interpreted the division as indicative of two migrations into the New World. (See, e.g., Neves, W. A., and M. Hubbe. 2005. “Cranial Morphology of Early Americans from Lagoa Santa, Brazil: Implications for the Settlement of the New World,” Proceedings of the National Academy of Sciences 102, 18309-18314). Instead, they observed a wide range of cranial variation which is independent of both chronology and geography (see below, Fig. 2).
The authors further note (p. 290) that
“…Considering the CI values as well as the geographical origin of the samples, and due to head length contribution, the skulls may be classified as dolichocephalic. This finding is consistent with he classical argument of South American physical anthropologists about the dolicocephalic morphology of most ancient skulls compared to more recent ones belonging to agricultural societies. However, when the shape and size components of cranial form are partitioned, the main “cephalic types” of classical physical anthropology are all well represented in the morphometric shape space (i.e. dolicocephalic, mesocephalic and brachicephalic ones), clustering independently of the chronological origin of the samples.”
The two morphological types construed by Walter Neves, Joseph Powell and other paleobiologists have always been hard to reconcile with the main finding from American Indian haploid genetics in the 1990s, namely that all principal mtDNA and Y-DNA types are evenly distributed across South, Central and North America (Merriwether, D. A., F. Rothhammer, and R. E. Ferrell. 1995. “Distribution of the Four Founding Lineage Haplotypes in Native Americans Suggests a Single Wave of Migration for the New World,” Am J Phys Anthropology 98 (4): 411-430). Ancient mtDNA obtained from early Holocene skulls failed to show stark molecular contrast between ancient and modern samples but confirmed the heterogeneity of ancient samples (Kemp B. M,. R. S. Malhi, J. McDonough, D. A. Bolnick, J. A. Eshleman. (2007) “Genetic Analysis of Early Holocene Skeletal Remains from Alaska and Its Implications for the Settlement of the Americas,” Am J Phys Anthropology 132: 605-621).
Proponents of the dual-migration theory acknowledge that theirs is not the only possible explanation of the observed split between ancient and recent American Indian skulls.
“This, however, does not completely exclude the possibility that the observed morphological diversity in America is the result of diachronic trends of differentiation, or progressive losses of the original variability present in the mother-population of Native Americans, especially if strong diversifying selection acted upon the morphological pattern brought into the continent by its first populations.” (Hubbe, M., W. A. Neves, K. Harvati. 2010. “Testing Evolutionary and Dispersion Scenarios for the Settlement of the New World,” PLoS ONE 5 (6): e11105.
Manriquez et al.’s data is consistent with these alternatives to the two-migration model. Pursuant of their data, they endorsed a single-wave model for the peopling of the Americas. However, this is not a throwback to the times when American Indian variation was perceived as unambiguously restricted and falling into a “Mongoloid” cluster. The reality behind the two divergent schools of thought on American Indian craniological variation, and this has largely come to light in the 1990s-2000s, is that American Indian skulls are exceptionally diverse, and that the “Mongoloid” type is just a subset of this broad variation found in the Americas. One should recall Peter Brown’s observation that “at present the earliest people with a generalised East Asian cranial morphology are probably found in the Americas. Is it a possibility that migration across the Bearing Straits went in two directions and the first morphological Mongoloids evolved in the Americas?”


I must say that the persistence of A, B, C and D throughout the Americas is very puzzling, as is the evidence of homogeneity expressed in this paper. What puzzles me are the clear cultural differences between essentially all groups north of Mexico and certain ones in Mexico and south of Mexico. This is very clear from the musical evidence. There are also striking similarities between the music of many of these southern groups and Melanesia, and many of these same groups are morphologically very different from what we see in the North, more squat and muscular, often resembling Melanesians.
This is a problem for me because the clear musical and morphological distinctions in the Americas don’t seem to correlate with genetic distinctions and I wonder why.
There are two genetic facts that, IMO, represent possible analogs of the cultural similarities between Africa, PNG and Amazonia.
1. mtDNA 9-bp deletion may be a relic that arose only once in human history. I blogged about it at http://anthropogenesis.kinshipstudies.org/2012/03/on-the-origin-of-mtdna-haplogroup-b-in-the-americas-9-bp-deletion-in-america-asia-and-africa/ and at http://anthropogenesis.kinshipstudies.org/2012/07/the-western-stemmed-tradition-clovis-and-mtdna-9-bp-deletion/
If this is so, then 9bp deletion in Pygmies and 9 bp deletion in Circumpacific will be survivals from the primitive state. The lack of 9bp deletion in PNG but the presence of a closely related hg P in PNG and Australia is explained as the loss of 9bp deletion in PNG. (There is 1 case of 9bp deletion in Australia.)
2. “Denisovan admixture” is found at highest frequencies among Papuans followed by South American Indians. http://anthropogenesis.kinshipstudies.org/2012/03/american-indians-neanderthals-and-denisovans-pca-views/
I don’t think that Denisovan “admixture” is “admixture” but rather phylogenetic archaisms that survived in PNG and the New World (also areas with highest linguistic diversity) but got lost in Africa.
German, are you aware of the following paper, dating from 1961?
http://www.sciencemag.org/content/134/3485/1077.abstract
One of the co-authors is Johannes Wilbert, with whom I studied that same year (or possibly the following year) at UCLA. I’m sure you know who he is, but you may not be aware of his pioneering genetic research, which predates Cavalli-Sforza’s and isn’t mentioned in his book. Are you aware of any of this blood type based research and do you think it’s been completely superseded by DNA research or would still have something meaningful to contribute?
I’m wondering whether anyone ever followed up on Wilbert’s research on the Diego gene.
If you are looking for a primordial population in the Americas, then absence of the Diego gene could be significant. And you’ll be interested to know that Warao shamans sing in “canonic-echoic” style.
Thank you, Victor. I wasn’t aware of Wilbert’s paper (although I know his other works, are you in touch with him, he’s still in UCLA, right?). I’ll take a look. I do know of the pre-Cavalli tradition of genetic research among South American Indians (James Neel, Rick Ward, Francisco Salzano etc.) and use it as an argument against out of Africa. These people argued that South American Indian tribes harbor extensive genetic and biological diversity comparable in volume to that of a cosmopolitan city. James Neel published a paper in Portuguese (Neel, James. “Estrutura populacional de amerindios e algumas interpretacoes sobre evolucao humana,” Origens, Adaptacoes e Diversidade Biologica do Homem Nativo da Amazonia. 1991) questioning the basic assumption of out-of-Africa, namely the panmictic model and serial founder effect arguing instead that the earliest human demes must have been heavily structured. I think that intellectual “lineage” has died out (the last paper down this venue was Ward’s Extensive Mitochondrial Diversity in an American Indian tribe http://www.ncbi.nlm.nih.gov/pmc/articles/PMC52581/) and merged with the Cavalli school of global DNA studies.
Thanks again for bringing it up.
I would offer one correction: mtDNA haplogroups are not “evenly” distributed throughout the Americas and I highly doubt Merriwether et al. (2005) would have described them as such. In fact, they mentioned two clines in the first line of their abstract! With regards to the presence of all 4 main haplogroups in all three of Greenberg et al’s linguistic groups, this is not really true. Northern Athabaskans and Inuit mainly have haplogroup A sequences. Southern Athabascans are known to be admixed.
The haplogroups of Europe are much more “evenly” distributed than those of the Americas.
If some of the earliest migrations into the Americas were climate-motivated, then they may have occurred around the same time.
“the two-way MANOVA for landmark data, showed significant differences in skull shape variation due to geographical origin …, but not to chronological period…” [data snipped for clarity]
You are right, Merriwether never used the term “evenly.” This is my shortcut to the following quote from Merriwether & Ferrell 1996 “The Four Founding Lineage Hypothesis for the New World: A Critical Reevaluation”, p. 245: ” …Many populations have all four founding lineage types (a number even have all of the subtypes) and that all types can be found in North, Central and South America. Multiple lineages are found in all three putative waves of migration (Amerind, Na-Dene and Eskaleut). We believe that this distribution is most consistent with a single wave of migration.”
I agree with all of your caveats, Gisele, but I do want to highlight the fact that, fundamentally, all major divisions of New World populations share widely divergent lineages belonging to mhg N and mhg M (mtDNA) and hg Q and hg C (Y-DNA). A good example is the presence of hg C in Eskimos, Na-Dene, North American Indians and, as of lately, Northwest South American Indians. This likely means that American Indian populations are very subdivided, or diverse on the tribal level (this is consistent with all the pre-Cavalli research in the Americas done by the likes of Rick Ward, Francisco Salzano and James Neel) and have been stable and subdivided for a very long time.
Sometimes, genetic drift results in the loss of some variants in some tribes and regions, but this doesn’t affect the whole picture. For instance, Na-Dene are almost all hg A, right, but it’s the same A as further down south, which suggests that they used to have C, D and B but lost them as a result of their conquest of the Subarctic. BTW, I’m not 100% convinced that Southern Athabascans got all of the non-A lineages from Pueblo. They have hg X, which is related to hg X in Algonquins. In “The Genius of Kinship,” I tackled South Athabascan kinship systems and came to the conclusion that they more faithfully reflect proto-Athabascan kinship than California or Northern Athabascan kinship systems. The latter, indeed, got transformed as a result of the conquest of the Subarctic. Southern Athabascan kinship patterns look much less modified, as if they avoided the demographic pressures associated with the expansion into the Subarctic or California. And this has nothing to do with Pueblo influence. I don’t think Southern Athabascans have always been where they are attested historically, but they either managed to slide down the Northwest Coast and remain intact, or Na-Dene originated further south from their putative homeland in Alaska or the northern Northwest Coast, e.g. somewhere in the Plateau area.
Also, while we are at it, I looked up Merriwether for hg B in Eskimos, and Merriwether & Ferrell 1996, p. 244, Table 2, report B1 and B2 (maybe an old notation) from Old Harbor Eskimos.
German, your references are all very old. You do know that several mtDNA articles on the Peopling of the New World have been published every year since 1995? 🙂
You wrote:
“Na-Dene are almost all hg A, right, but it’s the same A as further down south”
Yes, so? One possibility is that haplogroup A was carried over the landbridge and haplogroups B, C and D (not necessarily together) were brought via a more coastal route.
Finding 2-4 haplogroups together in most populations after 15,000 or so years – well what do you expect? Languages were not barriers; nor are they in the present.
I think the majority of geneticists agree that the southern Athabascans only brought A2 and a few D2 sequences down south. This is not just my opinion. You will find this interpretation in more recent journals.
The plateau is the most northern part of the predominantly haplogroup B region (or B and C) of the South west.
The Navajo frequency of haplogroup X is 1/167 and Apache 1/187; nothing to get excited about.
Gisele, I study all publications, old and new. But if hg B in Eskimos was reported in a 1996 paper it doesn’t make it false. I know all the recent studies on Athabascan migrations. Low frequency of hg X in Southern Athabascans is normal for such a low frequency haplogroup as X. But it indicates, rather unambiguously, that Athabascans, via Southern Athabascans, belong with “Amerinds,” not with Siberians, as a three-wave theory would want to have it. (This would make Kets migrate from North America, not the other way around.) This also means they originated further down south, not in the Subarctic where hg X is absent. Finally, it also means that the ancestors of Athabascans likely had other typical American Indian haplogroups, which means that haplogroups are rather evenly distributed in the Americas and the gaps, when they are found, come from more recent demographic processes within the Americas and don’t indicate separate migrations to the New World.
We may want to revisit recent papers devoted to Athabascan origins and see if Southern Athabascan hg B is indeed from Pueblo and not a native Na-Dene haplogroup coming from an earlier Plateau habitation, which as you write is part of the Greater Southwest range of hg B.
“…if hg B in Eskimos was reported in a 1996 paper it doesn’t make it false.”
Not false but Merriwether et al. might have been a little less confident about stating that haplogroup B sequences were present in all three of Greenberg’s linguistic divisions if they knew that no other haplogroup B sequences would be found in another 975 Eskimo and Aleut sequences (including 100 ancient). Frequencies of less than 1% should not be statistically significant for a language group.
Raff (2010) reported 2 haplogroup B HVR I lineages in their ancient Brooks River sample (938–1318 BP). While the variants reported definitely belong to haplogroup B, they were not determined to belong to the Native American clade; they were assumed to be:
“The most unexpected result from these analyses, however, is the presence of two HVRI sequences consistent with mtDNA haplogroup B2. These two samples share transitions at np 16189C and np 16217C that are diagnostic for haplogroup B2 (lineages B2 and B4 cannot be distinguished with HVRI, but B2 is the expected American founding haplogroup and we infer it here).”
The X sequences of the southern Athabaskans could easily have been acquired via admixture and so I wouldn’t consider them to be the best evidence of their connection with other Native Americans. The best evidence is that northern and southern Athabaskan mtDNA lineages have New World specific mutations shared with almost all other Native Americans. An exception is a couple of D2 sequences which clearly illustrate their connection with the north as there are no southern populations they could have acquired them from.
With regards to the Kets migrating from North America, I tried to prove that for a while but it is extremely difficult when researchers in other disciplines are interpreting correlations as due to movements in the opposite direction. I suspect you know something about that. 🙂 The Selkups have a couple of ‘A’ sequences of Native American origin (Tamm, 2007) and some Yeniseian artifacts in Russian journals have been described as “Americanoid” (Kozintsev, 1999).
The southern Athabaskans’ haplogroup A sequences have affinities with those of the north. Their ‘B’ sequences, on the other hand, have affinities with those of the south (Pima, Zuni, etc.). See Malhi (2008, Distribution of Y chromosomes among Native North Americans: A study of Athapaskan population history) who go into this in greater detail. They also state that the distributions of albumins Naskapi and Mexico are consistent with this interpretation. One interpretation in this article appears to be influenced by Vajda’s linguistic evidence: “This suggests that the majority of non-C Y chromosomes in the Navajo and Apache were contributed by non-Athapaskan populations in the Southwest, which mirrors the presence in the Apachean of mtDNA lineages belonging to haplogroups B and C.” (I don’t think so).
The NRY C sequences were the male counterpart of the Aleut/Saqqaq-like D2, IMO. Like D2, they have also been reported in Greenland.
The Athabascan case is fascinating. A couple of points:
1. The diffusion of hg X into southern Athabascans (from, say, Algonquins) is not an easy solve. If Southern Athabascans absorbed it from Algonquins, they would’ve got more frequent markers from them as well. But they didn’t. Hence, I’d still consider it a weak but suggestive indicator of the common origin between Na-Dene and other Amerindians complementing hg A2. Also Albumin Naskapi is product of common descent between Athabascans and Algonquins (as Smith et al. 2000 argued, see http://www.ncbi.nlm.nih.gov/pubmed/10727973). Southern Athabascans picked up Albumin Mexico from Pueblo.
2. If Na-Dene belong with other Amerindians, then the “Siberian” autosomal component detected in them by Reich et al. 2012 can’t be their original one and the First American component derived from admixture with other Amerindians. It means what Reich et al. 2012 found in Na-Dene are incipient Siberian lineages that were carried out back to the Old World and expanded there. Autosomes can’t be in direct contradiction with the haploids.
3. mtDNA D2 is a curious one. How could it have been brought from South Siberia to the New World AFTER the initial peopling (Tamm et al. 2007 http://www.plosone.org/article/info:doi/10.1371/journal.pone.0000829) as a separate migration that didn’t carry with it any other lineages attested in Tuvinians or Buryats? In addition, this one lineage somehow got into Na-Dene through gene flow but then did NOT get lost in Apache who supposedly went through a massive mtDNA bottleneck. D2 is a separate lineage in Na-Dene (16092), which is different from D2 in Eskimos, hence it couldn’t be the result of gene flow from Eskimos.
We need to step back and re-assess what was going on at the Pleistocene-Holocene boundary between the Old and New World. I think there was some significant lineage loss going on in northern North America. I think hg X, C and D2 were part of the proto-Na-Dene gene pool in addition to hg A2. Hg B could still be original in Na-Dene, too, but then some Southwestern versions were added in Southern Athabascans on top of the native Na-Dene Bs.
Between Merriwether and Raff, I think we can consider it established that Eskimos had hg B, which makes their original mtDNA pool also more diverse than the present one. Again, consistent with the finding of Y-DNA hg C in them.
Hg D2 must be original to the New World. It evolved in proto-Eskimos and proto-Na-Dene and then expanded into Northeast Asia and trickled down to South Siberia. If there are some phylogenetic problems with it, we need to entertain 2 options – 1) loss of some early D4 lineages in the New World; 2) a special, more divergent phylogenetic position of D2 within D4.
Regarding Kets, you and I need to bring it home to people that, all things being equal, it’s more likely that Kets came from the New World than the other way around. In The Genius of Kinship, I suggested a straigtforward derivation of a part of Ket kinship system from Na-Dene. The opposite process is not valid cross-linguistically.
Have you seen The Dene-Yeniseian Connection, ed. by J. Kari and B. Potter (2010)? In it, some scholars entertain a back-migration of Kets from Alaska.
One thing we need to watch out for is the possibility that what Vajda found was a trace of a much older and broader kinship that involved other (possible extinct) languages in America and Siberia, rather than a point-to-point connection between Na-Dene and Ket only suggesting a relatively recent (within 10,000 years) migration. Although linguistic method doesn’t usually stretch that far, Na-Dene conservatism is well known and who knows how far deep in time it stretches.
Also, they reported mtDNA hg W in Kets, which is sister to hg X, which I believe is native in Southern Athabascans.
I will respond to your e-mail shortly, too.
With regards to:
“It means what Reich et al. 2012 found in Na-Dene are incipient Siberian lineages that were carried out back to the Old World and expanded there. Autosomes can’t be in direct contradiction with the haploids.”
They are not (except, perhaps, concerning the number of recent migrations from Asia but see below):
mtDNA: D2 + D3 Asia –> northern North America
Y chromosome C : Asia –> northern North America (and a few more places)
Autosomal “Siberian Component”: Asia –> northern North America
“However, speakers of Eskimo–Aleut languages from
the Arctic inherit almost half their ancestry from a second stream of Asian gene flow, and the Na-Dene-speaking Chipewyan from Canada inherit roughly one-tenth of their ancestry from a third stream.” (Reich et al. 2012)
“We cannot confirm that Saqqaq derive from a stream of Asian gene flow distinct from that
which led to Na-Dene speakers like the Chipewyan.” (from Reich supplementary file)
Saqqaq mtDNA sequence belonged to D2.
Archaeological findings:
“In 2008 we used permafrost-preserved hair from one of the earliest individuals that settled in the New World Arctic (northern Alaska, Canada and Greenland) belonging to the Saqqaq Culture (a component of the Arctic Small Tool tradition; approximately 4,750–2,500 14C years before present (yr BP))14,15 to generate the first complete ancient
human mitochondrial DNA (mtDNA) genome.” (Rasmussen et al. 2010)
“The true Arctic coast was not colonized until after 4500 BP with the sudden appearance of the Arctic Small Tool tradition. Discovered in Alaska in 1948 as the Denbigh Flint Complex, makers of diminutive bipointed arrow points, of delicate bifacial implements for sidehafting in projectiles or knife handles, and of burins and microblades, by 4500 BP had covered the Arctic from Alaska to Greenland; in the east the Denbigh relatives were known as Pre-Dorset.” (Don E. Dummond)
Aleuts mtDNA sequences are about 60% D2.
Based upon complete sequence data (as it is not as easy to identify the sub-groups of D2 based upon HVR I mutations):
D2a – Aleut
D2a1 – Aleuts, Sireniki Eskimo, Saqqaq, Siberian Eskimo, Navajo?
D2a2 – Sireniki Eskimo, Chukchi, Eskimo
D2b1 – Buryat, Khamnigan, Yakut, Evenk, Tibet, Kalmyk
D2b2 – Bargut, Evenk
D2c – Buryat
D2 is a subset of Asian D4e, has a restricted distribution in the New World and is not very diverse.
No Apaches have been determined to have D2 mtDNA sequences to my knowledge; only Navajo at low frequency.
I interpreted Derbeneva 1992 to mean that they found D2a in an Apache individual. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC379174/
Per Reich, Eskimos, Na-Dene and Paleoasiatic peoples have the “First American” component. It’s unlikely that three independent waves of migration to the New World and Beringia received the “First American” component through admixture. It just makes much more sense that the two “Siberian” components originated in the New World off of the “First American” component.
The fact that “D2 is a subset of Asian D4e, has a restricted distribution in the New World and is not very diverse” only means lineage extinction in the New World (smaller populations) vs. higher lineage retention rate in Siberia (larger populations). The fact that D4h3 is also found in the New World supports the lineage extinction hypothesis because otherwise you need to postulate another migration to the New World to explain that very lineage. Again, it’s teleological to argue that D2 and only D2 was brought to Beringia by relatives of the Buryats and then D4h was brought by the relatives of Ulchi and Japanese as another migration. But no other D4 lineages came along in both cases and no other haplogroups, although D4 is so diverse in the NE Asia and there are many other, older, more diverse haplogroups in Asia that could’ve been brought to the New World but for some reason weren’t. Not to mention the fact that, for hg X, you’ll have to postulate a special migration from the Near East, again carrying only one selected haplotype.
I would also look very closely into the phylogeny. I wouldn’t be surprised if hg D2, for instance, will turn out to form the earliest split in the D4 phylogeny. For instance, G16129A is a “back mutation” shared by both D2 and D4a. When people built phylogenies they apriori assume that the migrations went mostly into the New World. Looking it in the other direction may end up being a worthwhile exercise.
Also, if you have been following Dienekes’s followups on Reich’s finding that non-Arctic Amerindians exhibit strong autosomal proximity to West Eurasians with gene flow, per Dienekes, going east to west (from the New World), you can get a bigger picture into which I’m trying to fit the more granular but also more unstable data from the haploids.
http://dienekes.blogspot.com/2012/08/east-eurasian-like-ancestry-in-northern.html
North West Europeans do have Asian admixture. I think it is mainly haplogroups D and Z in mtDNAs.
24/24 Karitiana Indians had mtDNA haplogroup D1 sequences. Likely they have a high frequency of NRY Q which would make them closer to NW Europeans in autosomal DNA than Siberians having non-Q sequences. That’s my take.
This is good information, thanks Gisele. The “Amerindian” component in Europe is more frequent in northern Europeans but it’s found in southern Europeans, too, where there are no D and Z lineages. My hypothesis is that ancient DNA from Upper Paleoilthic sites in Europe will identify pre-U haplogroups related to East Asian and Amerindian ones. There seems to be some evidence that Gravettian mandibulae had shovel-shaped incisors, which is an Asian-Amerindian dental trait.
Sorry, that should have been *NE* Europeans – mtDNA D and Z admixture due to the movements of Uralic populations. The D sequences are mainly D5 but there could be some D4. No Amerindian D1…
Then in order to reconcile autosomes and mtDNA we may need to postulate that D1 is the most divergent D clade and D5 is one of the later ones.
Also, D2 has been reported from Uto-Aztecan and other groups in Mexico: Pen ̃aloza-espinosa. Characterization of mtDNA Haplogroups in 14 Mexican Indigenous Populations, 2007.http://www.ncbi.nlm.nih.gov/sites/entrez?Db=pubmed&Cmd=ShowDetailView&TermToSearch=18078204
I don’t see any reference to this paper in PhyloTree. Could it be that they mistyped D4h3 as D2? Or is it a legitimate D2?
I do not think I have that particular paper but, based upon the following statement from the abstract, I can tell it does not refer to what is currently recognized as “D2”.
“Our data show that 98.6% of the mtDNA was distributed in haplogroups A1, A2, B1, B2, C1, C2, D1, and D2.”
Okay, I think I can add some clarity to this enigmatic hg D2 in Mexico. It must be indeed the same D2 as in Beringia or it’s its sister clade D4e1c reported by Kumar in Mexico (http://www.biomedcentral.com/1471-2148/11/293). It’s on PhyloTree now, too. In any case, in the light of this data, D2 in Southern Athabascans, if it’s real, doesn’t need to be a sign of their northern origin, as closely related lineages are found in Mexico. Also, in light of D4e1c, the hypothesis of a separate migration from South Siberia to Beringia becomes problematic.
On a broader level, American Indians are rich in D1 but they also have unique lineages in D4. These are the two biggest D clades. Only D4 and not D1 are found in Asia. This suggests that there was a migration out of the New World, and not the other way around. Better sampling in the New World will continue to turn up D4 lineages that ended up riding to higher frequencies in Asia.
The date of Derbeneva’s paper should be 2002. 🙂 If she wrote that an Apache of Torroni (1993) was later determined to belong to D2, then she must be right.
The reference for the Navajo D2 is Budowle et al. (2002) but they just classiifed the two sequences as “D” in their supplementary files.
With regards to:
“Looking it in the other direction may end up being a worthwhile exercise.”
German, despite all of the other haplogroups the Japanese and Koreans have their frequency of haplogroup D sequences (many kinds) is still somewhere around 35%. Need I say more? Is it more likely that rare haplogroups of Asian populations were reintroduced to the New World or ones that are frequent in populations nearby?
If rarity, restricted distribution, low diversity, belonging to the phylogeny of another continent, etc. does not signify admixture to you, what does?
You wrote:
“Have you seen The Dene-Yeniseian Connection, ed. by J. Kari and B. Potter (2010)? In it, some scholars entertain a back-migration of Kets from Alaska.”
No, I have not read it. I would buy a copy if it was available in electronic (pdf) format (even though I would not be able to follow the large linguistic parts)
An excerpt from one of several book reviews:
[Paper13] Yeniseian: Siberian Intruder or Remnant? (Michael Fortescue) (p.310-315). In this paper MF explains how he has moved from sceptical to moderately supportive of the ND-YE connection. He examines four scenarios of split and dispersal of this “family” with a focus on the idea that YE would be closer to Tlingit than to the rest of ND. One of the scenarios include a return from North America to Siberia.
http://diachronica.pagesperso-orange.fr/TMCJ_vol_2.1_Fournet_Review_of_APUA5_2010.pdf
The mention of the Tlingit is interesting because the two Selkup A2a sequences are closest to those of 2 Aleuts and 1 Tlingit. [A2a sequences are carried by northern & southern Athabaskans, Aleuts and Inuit.]
Based upon other mtDNA similarities, the Selkups and Kets appear to have intermixed. It happens, German.
With regards to the haplogroup W, the Kets have a high frequency of European admixture; more sequences of western Eurasian origin than of Asian, in fact.
I just wrote a post on Dene-Yeniseian with video links to most recent talks by Vajda and others. Let me know what you think.
Your argument against hg W being original to Ket makes sense.
Kumar (2011) wrote:
“After removing the related individuals from our maternal lines and those of European or African
descent….”
What about Asian? We have Asian Americans here too.
I have reason to doubt that Kumar’s two D4e1c sequences are “Native American”. This is because a similar sequence was obtained by Derenko (2010) which has not been added to the Phylotree yet. It was obtained from a Czech… who had Asian maternal ancestry.
Both the “Czech” and 1 of the Mexican have coding region mutation 6366A & 16188T in common and . As soon as additional D4e1c sequences are obtained, the 6366A will become the defining mutation for this sub-group rather than 14207. It looks like Kumar et al. missed the 6366A on one sequence as the Czech does not have 14207.
Okay, this is a fair push. Let’s wait. The D2 in Mexico still remain a mystery, though. But since it has been reported I would be cautious about the whole D2 migration from South Siberia to Beringia and into South Athabascans.
I have D4e1 haplogroup but my mother is from Austria near Tyrol. I have 6366A and 16188T but not 14207. We have no “known” Asian ancestry.
Like the Czech…. I would place some sequences of SE China/Taiwan next closest to those of this small group (rather than the NE Asian D2e sequences).