Nivkh and Chukotko-Kamchatkan Linguistic Relationship and Its Genetic Correlates
Lingua Vol. 8, Issue 121, June 2011, 1359-1376 http://dx.doi.org/10.1016/j.lingua.2011.03.001
The Relationship of Nivkh to Chukotko-Kamchatkan Revisited
Michael Fortescue
With the availability today of reliable materials for comparing the languages that in the past have been lumped together under the rubric ‘‘Paleosiberian’’ it has become possible to reassess the genetic relationship – or lack of it – between the individual languages of this traditional grouping. It will be demonstrated that the case for the genetic relationship of two of the constituent groups, Chukotko-Kamchatkan and the isolated Amuric language Nivkh (Gilyak), is actually quite strong, although the rest of the grouping must indeed be abandoned as a genetic unit. A case is made for reconstructing a Chukokto-Kamchatkan- Amuric proto-language associated with the Neolithic of the Lower Amur and adjacent coasts of the Sea of Okhotsk. Emphasis is laid especially on the morphology and shared typological features of these languages, but numerous lexical items based on systematic sound correspondences are also introduced. A plausible archaeological framework elucidating the evident closeness of the two linguistic entities is also sketched.
Michael Fortescue, a British-born linguist teaching at the University of Copenhagen, is famous for his intricate analyses of linguistic relationships (genealogical, typological and contact-based) on both sides of the Bering Strait and their possible archaeological and population genetic correlates. In 1998, they culminated in a dense volume entitled Language Relations Across The Bering Strait: Reappraising the Archaeological and Linguistic Evidence. One of the highlights of the book is the proposed kinship between Eskimo-Aleut, Chukotko-Kamchatkan (CK), Yukaghir and Uralic languages forming a new Uralo-Siberian family. This hypothesis was further confirmed by Uwe Seefloth (“Die Entstehung polypersonaler Paradigmen im Uralo-Siberischen,” Zentralasiatische Studien 30, 2000, 163-191) and is considered to be one of the more probable and respectable macrophyla. I corresponded with Michael Fortescue back in the early 2000s trying to organize an interdisciplinary conference on linguistic and cultural data pertaining to the prehistory of Beringia.
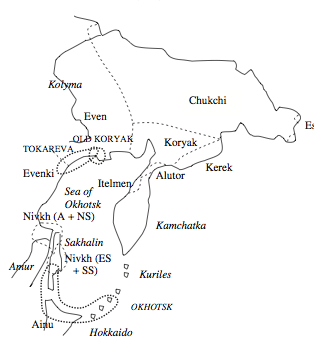 In the new article published in a mainstream linguistic journal, Fortescue revisits the greatly improved linguistic data from Northeast Asia and advances the hypothetical Chukotko-Kamchatkan-Amuric (CKA) language family encompassing Chukchi, Koryak and Itelmen, on the one hand, and Nivkh, the former isolate spoken by some 1000 people residing in easternmost Siberia in the basin of the Amgun and Amur rivers and on the northern half of Sakhalin, on the other. Evidence is typological, morphological and lexical. Morphological evidence looks especially solid and covers nominal and verbal morphology, pronouns, case systems, deverbal suffixes and numeral classifiers. Numeral classifiers are lacking in CK but are pervasive in Nivkh. Fortescue explains Nivkh classifiers as the areal feature also found in Korean and Japanese and unearths behind it in Nivkh the traces of a simple division of nouns into animate and kin vs. inanimate similar in form and function to the one found in CK.
In the new article published in a mainstream linguistic journal, Fortescue revisits the greatly improved linguistic data from Northeast Asia and advances the hypothetical Chukotko-Kamchatkan-Amuric (CKA) language family encompassing Chukchi, Koryak and Itelmen, on the one hand, and Nivkh, the former isolate spoken by some 1000 people residing in easternmost Siberia in the basin of the Amgun and Amur rivers and on the northern half of Sakhalin, on the other. Evidence is typological, morphological and lexical. Morphological evidence looks especially solid and covers nominal and verbal morphology, pronouns, case systems, deverbal suffixes and numeral classifiers. Numeral classifiers are lacking in CK but are pervasive in Nivkh. Fortescue explains Nivkh classifiers as the areal feature also found in Korean and Japanese and unearths behind it in Nivkh the traces of a simple division of nouns into animate and kin vs. inanimate similar in form and function to the one found in CK.
Fortescue is not one hundred percent sure that the evidence that he amassed proves common descent between CK and Nivkh. Ancient contacts taking place some 4,000 YA cannot be discounted, but at least he greatly reduced the chances that the similarities between CK and Nivkh are due to random convergence.
Taking a quick look at mtDNA profiles of CK- and Nivkh speakers, leads to the following observations (data from Jin, Han-Jun, Ki-Cheol Kim, and Wook Kim. “Genetic Diversity of Two Haploid Markers in the Udegey Population from Southeastern Siberia,” American Journal of Physical Anthropology 2010 142 (2): 303-313).
Chukchi (A 67.7%, C 12.3%, D 10.8%, G 9.2%)
Koryak (A 5.2%, C 36.1%, D 1.3%, G 41.9%, Y 9.7%, Z 5.8%)
Itelmen (A 6.4%, C 14.9%, D 0%, G 68.1%, Y 4.3%, Z 6.4%)
Nivkh (A 0%, C 0%, D 28.6%, G 5.4%, Y 66.1%, Z 0%)
Nivkhs lack hg A and C, which are prominent among CK populations (and probably derive from a back-migration from the Americas), but they compensate by having high frequencies of hg Y, which is rare among CK peoples and non-existent in the New World. Other Amur groups (Udegeys, Oroks, Ulchi and Negidals) are also rich in hg Y, which immediately suggests that Nivkhs absorbed hg Y from their Tungusic-speaking neighbors after the breakdown of the hypothetical CKA language family. The loss of hgs A and C may have occurred through drift as Nivkhs show lower gene, nucleotide and haplotype diversity than the CK-speakers.
Y-DNA shows a similar dynamic between CK and Nivkhs. Both sets of populations have high frequencies of hg C3 (for Nivkhs, see Tajima, Atsushi, et al. “Genetic Origins of the Ainu Inferred from Combined DNA Analyses of Maternal and Paternal Lineages,” J Hum Genet 2004, 49, 187-193) but Nivkhs are also 28.6% hg O. CK, just like New World populations, do not have hg O. At the same time, Nivkhs do not seem to have hg Q, which in CK populations has, again, New World origin. Again, the hypothesis is that Nivkhs migrated from the north and entered the high O-zone. They absorbed hg O through admixture after their genetic tie with CK had been severed.
Both mtDNA and Y-DNA, therefore, paint the same picture of Nivkhs being intrusive in the Amur region, absorbing “southern” mtDNA and Y-DNA lineages are losing some of the “northern,” Paleoasiatic, Amerindian-derived markers. Correspondingly, on the linguistic side, per Fortescue, they borrowed numeral classifiers, which are very common in the languages of Southeast Asia, from a “southern” source. Fortescue (2011, 17), however, tentatively associates proto-CKA with Tokareva Culture of the mid-northern Sea of Okhotsk coast dated at about 3500 BP and derives it from the Lower Amur. Genetic evidence seems to suggest a reverse scenario.
Although geneticists (see, e.g., Schurr, Theodore, et al. 2000. “Mitochondrial DNA Diversity in Lower Amur River Populations, and Its Implications for the Genetic History of the North Pacific and the New World,” Am J Phys Anthropology Suppl. 30: 274-275) have been arguing against a special connection between Nivkhs, on the one hand, and Paleoasiatic populations, on the other, there seem to be good reasons to believe that the gap between the two sets of populations was created as a result of gene flow into Nivkhs from the original inhabitants of the Amur River basin. In fact, Fortescue’s linguistic argument makes perfect sense genetically once we peel off the sheer veneer of “southern” admixture (mtDNA hg Y and Y-DNA hg O, ) from Nivkhs and allow for some original lineage loss (mtDNA hg A and C, Y-DNA hg Q) in Nivkhs. On the basis of cultural, linguistic and distributional evidence, Franz Boas (“The Jesup North Pacific Expedition,” in Proceedings of the 13th International Congress of Americanists. Easton, PA: Eschenbach, 1905, 91-100; “The History of the American Race,” Annals of the American Academy of Sciences 21, 1912, 177–183) formulated a hypothesis whereby Chukchi, Koryak, Yukaghir, Itelmens (Kamchadals) and Nivkhs represent back-migration from the New World (see my out-of-America III) separated from its source by intrusive Eskimo-Aleut-speakers coming from Central Canada. (He did not bring up Kets, which now constitute another good candidate for a back-migration from the Americas, again on the strength of both linguistic and genetic evidence.) Boas dated this back-migration to the end of the last Ice Age, which makes proto-CKA likely older than Fortescue’s 4,000 YA. The Boasian “back-migration,” or “Eskimo wedge” scenario remains a viable explanation of the genetic and linguistic pattern in Northeast Asia and Beringia.


Interesting topic. It is possible to fine tune the haplogroup frequencies similar to the following and tease out the connections with northern North Americans, East Asians, SE Asians (and even Ket i.e. A8).
Chukchi
Haplogroup A2* 3
Haplogroup A2a 44
Haplogroup A2b 21
Haplogroup C4b2 5
Haplogroup C5a2 5
Haplogroup C4* 1
Haplogroup D3 3
Haplogroup D2 9
Haplogroup D4h1 1
Haplogroup D4o2 1
Haplogroup G1b 10
Total 103
Koryak
Haplogroup A2b 5
Haplogroup A2a 6
Haplogroup A8 9
Haplogroup A* 1
Haplogroup C5a 37
Haplogroup C4b2 44
Haplogroup C4a1 6
Haplogroup C4* 10
Haplogroup Z1a 13
Haplogroup D3 6
Haplogroup Y1a 21
Haplogroup G1b 99
Total 256
Itelmen
Haplogroup A8 3
Haplogroup C5a2 6
Haplogroup G1a 32
Haplogroup Y1a 2
Haplogroup Z1a 3
Total 46
Nivkhs
Haplogroup Y2b 5
Haplogroup Y1a 69
Haplogroup G1a 16
Haplogroup D4o2 5
Haplogroup D4o1 2
Haplogroup D4m2 15
Haplogroup D4a1e 1
Total 113
None of the above populations have the C1a haplogroup which would have been derived from the Americas. The path indicated was more to the south (Japan, Ulchi, Sakhalin Oroks, Buryats, Mongolia, Nanaitci, Altai). If you look back at the illustration you posted from Tamm et al., you will see that A2a and C1a migrations are shown separately. For some odd reason, they did not mention A2b in their article. Since practically all of the American and Greenlandic Inuit ‘A’ sequences belong to A2a and A2b, it seems likely that both were carried together.
The Ainu are also “rich” in haplogroup Y (20%) and phylogenetically related N9b (8%).
Haplogroup Y has both northern and SE Asian branches yet it is a relatively small haplogroup. It is possible that the Nivkhs have sequences belonging to both branches but I would like to see this confirmed via RFLP. See Macaulay 2005 (Single, Rapid Coastal Settlement of Asia Revealed by Analysis of Complete Mitochondrial Genomes). It appears that the nomenclature for Y has changed after publication of this article as the Y sequences of Sumatra definitely belong to Y2a1 (have 12161C 11299C like the sequences of the Philippines) and some northern Y2 complete sequences belong to Y2b (perhaps also a couple of the Nivkhi).
Has Fortescue compared the Nivkhi language to those of SE Asia? If you have a copy of his article, can you send it to me?
This is all great info, Gisele. Fortescue isn’t interested in SE Asian connections of Siberian languages. He’s very cautious and works one step at a time, plus he’s more of a specialist in North Pacific Rim languages. I’m not aware of any proposals linking Nivkh with SE Asian languages.
On the mtDNA side it’s surprising that Nivkh is as high as Nias in SE Asia in the frequency of such a small haplogroup. (See van Oven et al. Unexpected island effects at an extreme: reduced Y-chromosome and mitochondrial DNA diversity in Nias).
I did notice that mtDNA C1a is found in Ulchi and that this is interpreted as a trace of a back-migration from the New World. (Ulchi also have D1a).
In my heterodox mind, I look at the logic of haplogroup distributions differently. All these A8, A10, C5, D4, G, Z, Y etc. lineages must have formed and spread since the separation between Old World and New World populations, since they are not found in the Americas.
Ainu presumably picked their mtDNA C3 lineages from Nivkhs (see Tajima quoted in the post). I wonder if they got they mtDNA Ys from Nivkhs, too, although N9b is not found in Nivkhs.
No Asian haplogroup D sequences, to my knowledge, have New World-specific 2092T (which is the D1 coding region marker). Therefore, there is no evidence of the back-migration of this particular sequence type. However, some Asian D & G sequences are indistinguishable from D1 *in the first hypervariable region*. When the remaining portions of the sequences are amplified, there are fairly substantial differences between the Ulchi and Jomon D1 look-alikes and D1. In fact, the Ulchi/Jomon ‘D’ sequences which contain 16325C are closer to the ‘On Your Knees Cave’ D4h3 than to D1 based upon coding region mutations. Adachi (2011) corrected the classification provided in their 2009 article:
“In our previous study (Adachi et al., 2009), some of
the Hokkaido Jomon mtDNAs were assigned to the haplogroup formerly named as D1a (Starikovskaya et al., 2005) or D10 (Volodko et al., 2008). According to additional coding region analyses, this haplogroup is now renamed as haplogroup D4h2 (Van Oven and Kayser, 2009).
These articles are worth looking at, though, due to the Jomon N9b sequences which, as I wrote earlier, are phyogenetically related to the Ainu/Nivkhi Y. The Ulchi also have a substantial amount of ‘Y’ sequences.
The way that the Amur region differs from most other East Asian is the higher frequencies of sequences belonging to the N (xR) group of haplogroups; to which Asian/Native American ‘A’ belong. ‘N (xR) sequences did not derive from any M or R haplogroups but they eventually have to connect with SE Asian, Australian Aborigine N(xR) haplogroups and those of western Eurasia.
While the distribution of N9b would really help, these sequences are difficult to recognize via HVR I mutations alone.