Yuzhnyi Olenii Ostrov: Ancient mtDNA Evidence for Amerindian Admixture in Europe
Ph.D. dissertation, University of Adelaide, School of Earth and Environmental Sciences, Australian Centre for Ancient DNA Ecology and Evolutionary Biology, 2011
Mitochondrial DNA in Ancient Human Populations of Europe
Clio Der Sarkissian
Abstract. The distribution of human genetic variability is the result of thousand years of human evolutionary and population history. Geographical variation in the nonrecombining maternally inherited mitochondrial DNA has been studied in a wide array of modern populations in order to reconstruct the migrations that have participated in the spread of our ancestors on the planet. However, population genetic processes (e.g., replacement, genetic drift) can significantly bias the reconstruction and timing of past migratory and demographic events inferred from the analysis of modern-day marker distributions. This can lead to erroneous interpretations of ancient human population history, a problem that potentially could be circumvented by the direct assessment of genetic diversity in ancient humans. Despite important methodological problems associated with contamination and post-mortem degradation of ancient DNA, mitochondrial data have been previously obtained for a few spatially and temporally diverse European populations. Mitochondrial data revealed additional levels of complexity in the population history of Europeans that had remained unknown from the study of modern populations. This justifies the relevance of broadening the sampling of ancient mitochondrial DNA in both time and space. This study aims at filling gaps in the knowledge of the genetic history of eastern Europeans and of European genetic outliers, the Saami and the Sardinians. This study presents a significant extension to the knowledge of past human mitochondrial diversity. Ancient remains temporally-sampled from three groups of European populations have been examined: north east Europeans (200 – 8,000 years before present; N = 76), Iron Age Scythians of the Rostov area, Russia (2,300 – 2,600 years before present; N = 16), Bronze Age individuals of central Sardinia, Italy (3,200 – 3,400 years before present; N = 16). The genetic characterisation of these populations principally relied on sequencing of the mitochondrial control region and typing of single nucleotide polymorphisms in the coding region. Changes in mitochondrial DNA structure were tracked through time by comparing ancient and modern populations of Eurasia. Analysis of haplogroup data included principal component analysis, multidimensional scaling, fixation index computation and genetic distance mapping. Haplotypic data were compared by haplotype sharing analysis, phylogenetic networks, Analysis of the Molecular Variance and coalescent simulations. The sequencing of a whole mitochondrial genome in a north east European Mesolithic individual lead to defining a new branch within the human mitochondrial tree. This work presents direct evidence that Mesolithic eastern Europeans belonged to the same Palaeolithic/Mesolithic genetic background as central and northern Europeans. It was also shown that prehistoric eastern Europeans were the recipients of multiple migrations from the East in prehistory that had not been previously detected and/or timed on the basis of modern mtDNA data. Ancient DNA also provided insights in the genetic history of European genetic outliers; the Saami, whose ancestral population still remain unidentified, and the Sardinians, whose genetic differentiation is proposed to be the result of mating isolation since at least the Bronze Age. This study demonstrates the power of aDNA to reveal previously unknown population processes in the genetic history of modern Eurasians.
It’s sometimes the case that whole-genome analysis reveals molecular patterns that are indiscernible using haploid genetic systems such as mtDNA and Y-DNA. For instance, no evidence for “archaic admixture” (Neandertals and Denisovans) has been reported so far to match the whole-genome signatures thereof. The situation with the so-called “Amerindian admixture” in (West) Eurasia is similar: first detected by both professional geneticists and genome bloggers using STRUCTURE and ADMIXTURE analyses of autosomal markers, “Amerindian admixture” was further confirmed in a whole-genome study. But until recently neither mtDNA nor Y-DNA had anything to offer by way of corroborating the diploid data. The smaller effective population size of haploid systems, the small size of human populations in the pre-agropastoral age as well as the limited sampling of modern populations are the obvious explanations for the discrepancy. Ancient haploid DNA is the surest way to confirm or disprove the patterns of “admixture” detected in the whole-genome analyses of modern human populations.
Clio Der Sarkissian’s dissertation reports on ancient DNA from the Bronze Age Bolshoi Olenii Ostrov site (3500 YBP) in the Kola Peninsula (Kol’skii poluostrov, in Russian), the Early Neolithic Popov site (7000 YBP) and the largest Mesolithic burial ground in Europe – the Yuzhnyi Olenii Ostrov (Oleneostrovskii mogil’nik, in Russian) site in Karelia (northwestern Russia) dated at 7500-7000 YBP (see below).
Karelia has historically been occupied by Karelians (or Karely in Russian), the speakers of the West Finnic branch of the Uralic family, but the ancient DNA recovered from the site suggests that there was no strong continuity between Yuzhnyi Olenii Ostrov people and modern Karelians, Finns or Saami, hence Uralic-speakers must have colonized this area in post-Mesolithic times. Craniologically, the Yuzhnyi Olenii Ostrov burial is dominated by Caucasoid morphology (left) but, importantly, there is a small number of skulls that display Mongoloid traits (right). Odontologically, the burial shows elevated frequencies of several Sinodonty (Northeast Asian-Amerindian) traits, including shoveling (the same trend as observed in other places such as the Caucasus), six-cusp upper molar, deflecting wrinkle of the metaconid and distal trigonid crest, which can be interpreted as either the preservation of archaic (plesiomorphic) tooth morphology in Mesolithic Europeans (e.g., six-cusp UM is found as far back as Homo erectus) or as gene flow from Asians and or American Indians (see Зубова А.В. 2011. “Одонтологические данные к проблеме «монголоидности» населения Восточной Европы в мезолитическую эпоху,” Вестник Московского университета. Серия XXIII. Антропология. № 1).
Among the surprising findings from Yuzhnyi Olenii Ostrov mtDNA are hg H (a highly frequent modern European haplogroup, which so far has been poorly attested in pre-Neolithic sites) and, especially, hg C1, which is found at highest frequency and diversity in the New World. Hg U2e is also interesting as it’s related to hg U2 recovered from the earliest European mtDNA sample in Kostenki (southern Russia) coming from the 30,000 YBP time period. Hg U5b1 and hf V typical for modern Saami are not attested at Yuzhnyi Olenii Ostrov, but the sister clade of U5b1, U5a, is found in Yuzhnyi Olenii Ostrov, Bolshoi Olenii Ostrov and modern Saami. The sublineage attribution of of U5a in the former is unknown.
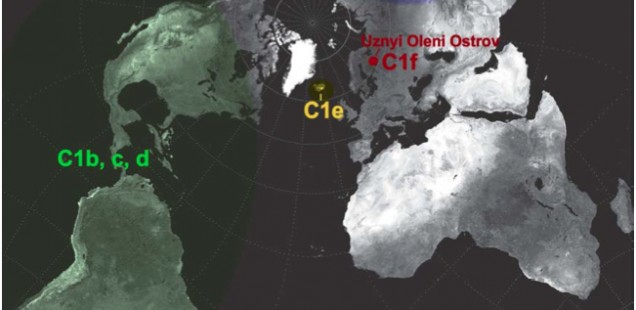
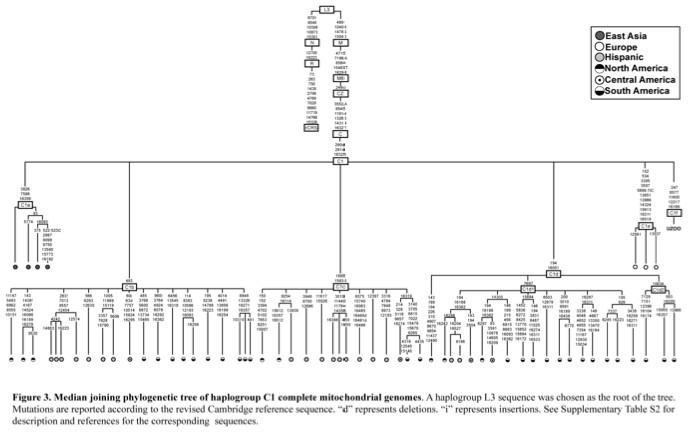
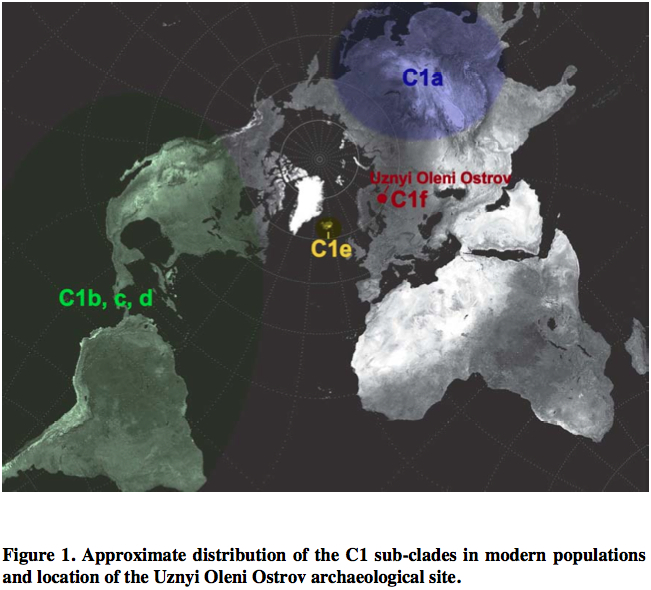
Most of world diversity in hg C1 is in the Americas. Hg C1 is known in the New World in three high-frequency forms – C1b, C1c and C1d (see below, from Der Sarkissian 2011, 165).
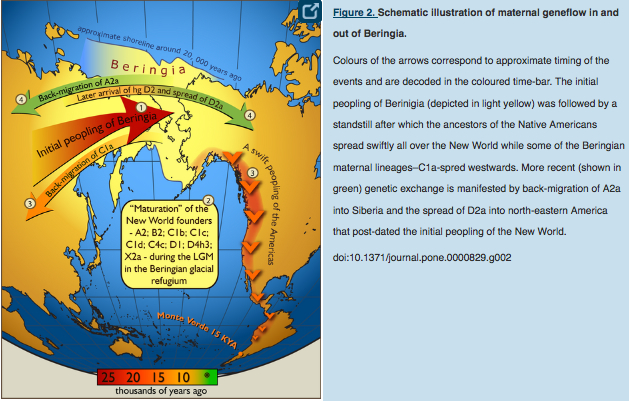
C1a is a rare lineage found in East Asia among Altaians, Oroks, Ulchi, Nanaitcy and Japanese. (Der Sarkissian, for some reason, attributes C1a to populations of Western Siberia, as in “The genetic affinity of Uznyi Oleni Ostrov/Popovo with Western Siberians is based on the presence of haplogroup C1,” p. 114) A back-migration from the America was hypothesized to explain its origin (see below, from Tamm et al. “Beringian Standstill and Spread of Native American Founders.” PLoS ONE 2 (9).)
Another Old World C1 lineage, namely C1e, has recently popped up not in Siberia or East Asia where American Indians are thought to have originated, but in Icelanders (Ebenesersdóttir et al. 2011. “A new subclade of mtDNA haplogroup C1 found in Icelanders: Evidence of pre-Columbian contact?” Am J Phys Anthropol 144, 92-99). (Comp. the occurrence of some Sinodonty dental traits in Icelanders.) Its phylogenetic proximity to the Amerindian clades and the physical proximity of Iceland to the New World via the Atlantic invoked recent (past 1000 years) admixture from a New World population to the Norse as an explanation. It’s important that neither in the case of C1a, nor in the case of C1e researchers hesitate to postulate an Amerindian origin for these Old World branches of C1. The recent origin of C1e in Iceland, however, can hardly withstand close scrutiny because hg C is not found in the Inuits with whom the Vikings might have interacted around 10th century AD. Additionally, C1 was reported in Germans, Canarians, Indians and Bashkirs (these sequences lack np 16356 HVR-I diagnostic of the sub-clades C1a and np 16051 HVR-I diagnostic of C1d) further reinforcing the impression that it’s an old European clade driven into obscurity by either selection or massive population replacements.
The discovery of a new clade of C1, C1f, in a Mesolithic site in northwestern Russia suggests that C1e is of ancient origin and connects Siberian C1a with Icelandic C1e to form a Circumpolar/Circumpacific chain of C1 attestations going back to at least the Early Holocene. Der Sarkisssian offers a nice map showing the breadth of the distribution of hg C1.
At the same time, Der Sarkissian’s strangely categorical claim (p. 168) that “the possibility of a prehistoric genetic influence from the Americas into Mesolithic Europe is highly unlikely” is a gross overstatement. Considering the fact that a whole-genome analysis corroborates the presence of Amerindian admixture in Europe (with northern Europe being more affected than southern Europe), hg C1a is already considered to be the product of a back-migration from the New World to eastern Siberia, craniologically Mongoloid morphology has the earliest appearance in North America, and not East Asia (Peter Brown (1999). “The First Modern East Asians? A Further Look at Upper Cave 101, Liujiang and Minatogawa,” in Interdisciplinary Perspectives on the Origins of the Japanese, International Research Center for Japanese Studies. Kyoto. Pp. 105-130. 1999) and archaeologically the quintessential Paleoindian fluted point industry is known to have expanded northward to Alaska and Northeast Asia, it’s not a stretch to imagine that some of these early “Mongoloid” groups migrated across Siberia into northern Europe at the end of the last Ice Age and merged with the pre-existing Caucasoid populations. The rarity of hg C1 in the ancient mtDNA samples in Europe may indicate a very targeted and restricted penetration of migrant forager groups that crossed Beringia after the melting of the glaciers. At the same time, Yuzhnyi Olenii Ostrov may be a refugium with deeper roots in European Upper Paleolithic, and hg C1 a relic surviving into the European Mesolithic from earlier times due to geographic isolation and long-term local endogamy reported for Yuzhnyi Olenii Ostrov by craniologists and odontologists (Jacobs K. (1992). “Human population differentiation in the peri-Baltic Mesolithic: The odontometrics of Oleneostrovskii mogilnik (Karelia),” Human Evolution 7 (4): 33-48).
Alternatively, Der Sarkissian (p. 168) is correct in noticing the similarity between the distribution of mtDNA hg X2 and C1.
“However, in the eventuality that further sampling of complete mtgenomes in the Americas reveals the presence of additional haplotypes belonging to C1f, it would suggest an evolutionary history similar to that of mtDNA Hg X2. Like Hg C1, Hg X2 displays relatively low frequencies and a broad distribution (in the Near East, Europe, Central Asia, Siberia as well as North America for clade X2a; Reidla et al., 2003). One model for the present distribution of Hg X2 suggests that the X2a clade split early from the rest of the X2 lineages in the Near East, and spread to reach east Siberia before participating in the second wave of migration into the Americas through admixture with Beringian populations (Perego et al., 2009). A similar scenario involving an early split of the different C1 clades in the Near East or Central Eurasia followed by their spread and isolated evolution could be considered as an explanation for the wide geographical distribution of Hg C1. However, this scenario currently lacks substantial support.”
She errs in assuming that the geographic distribution of X2 is continuous between America and Europe/Middle East: the meager number of X2 sequences detected in Siberia are considered to be products of a recent eastward migration of X2 from West to East Eurasia and not the retentions from the original dispersal of X2 between the New World and the Old World. But overall the similarity in distribution and frequency between hgs X2 and C1 is indeed remarkable. The finding of hg C1 in a Mesolithic sample suggests that the current absence of hg X2 from pre-Neolithic human remains in Europe may just as well be a function of limited sampling. The Paleolithic and Mesolithic genetic map of Europe must have been very different from the modern one: small pockets of geographically restricted lineages (H, C1, X2, U, etc.) were later overridden by Neolithic and Bronze Age population replacements.
Der Sarkissian makes an argument in favor of discontinuity between Mesolithic Europeans from Yuzhnyi Olenii Ostrov and modern West Finnic populations such as the Saami. This conclusion comes as a departure from the case built by odontologists (Jacobs 1992) that this burial “represents the earliest evidence for the establishment in the taiga zone of populations whose descendants are the modern Saami.” At the same time, there are clear similarities between the mtDNA pools of the Bronze Age Europeans from Bolshoi Olenii Ostrov and the present-day Saami such as the presence of East Siberian hg Z1, which entered the Saami mtDNA pool somewhere between the Mesolithic and the Bronze Age and, according to Der Sarkissian, likely with the spread of Kama culture.
“The presence of Z1 in samples from Bolshoy Oleni Ostrov establishes a direct genetic link between a Bronze Age population in NE-E and modern-day populations of the Volga-Ural region and Saami. The fact that Z1 was found in Bolshoy Oleni Ostrov but not in association with any Saami-specific haplotype further suggests that it must have had an independent origin and could have been integrated into the Saami gene pool via a separate migratory event” (Der Sarkissian 2011, 112).
The similarities between the modern Saami mtDNA gene pool and the Chalmny-Varre site from the 1800s are even stronger.
“At the haplogroup level, a genetic signature of the modern-day Saami is their high frequencies of haplogroups V and U5b (41.5% and 48% respectively; Ingman & Gyllensten, 2007) this was also observed in Chalmny-Varre: V (64%), U5b (29%), and U5a (7%). A haplotypic signature of the Saami, theU5b1b1 haplotype 16144C- 16189C-16270T, could also be detected in the population of Chalmny-Varre. This characteristic mutation motif has been coined the ‘Saami motif’ as it is very rare outside Saami groups but highly frequent among them all (Sajantila et al., 1995). Moreover, the particular V haplotypes sequenced in the population of Chalmny-Varre also show affinities with the present-day Saami, as the basal 16153A-16298C haplotype was reported in all the modern-day Saami populations sampled so far. Haplogroup U5a,which is present but less frequent in modern-day Saami populations (2.3%), was also detected in 3 individuals from Chalmny-Varre (7.1%)” (Der Sarkissian 2011, 106-107).
The only difference between Chalmny-Varre and modern Saami is the absence of hg Z1 among the former. This further indicates that ancient Saami gene pool was strongly structured due to the varying impact of East Siberian and West Eurasian admixture on the different Saami subpopulations.
The expansion of Kama culture (7000-6000 YBP) took place not too much later than Yuzhnyi Olenii Ostrov, hence it’s still possible that Yuzhnyi Olenii Ostrov represents the “original” mtDNA gene pool of the Saami, which later got diluted by East Siberian and West Eurasian genetic influences. This would preserve intact the theory that the Saami represent a remnant of a post-glacial re-expansion from the Franco-Cantabrian refugium along the western coast of Norway (see Tambets et al. 2004. “The Western and Eastern Roots of the Saami: The Story of Genetic “Outliers” Told by Mitochondrial DNA and Y Chromosomes,” Am J Hum Genet 74, 661-682). This would mean that ancient Saami possessed mtDNA hg C1, hence the striking cultural parallels between Saami and North American Indians (e.g., sauna and sweatlodge – the custom virtually absent in Siberia, see Lopatin I. A. “Origin of the Native American Steam Bath,” American Anthropologist 62 (6): 977-993) may finally have acquired a genetic foundation – the foundation watered down by subsequent gene flow from Siberia and West Eurasia into the Saami. Lopatin was so impressed with the similarities between sweatbaths among Saami and North American Indians that he hypothesized a direct connection between the former and the latter across the Atlantic – just like geneticists have entertained the possibility that hg C1 in Icelanders represents recent trans-Atlantic contacts. But in the light of the distribution of hg C1 (see above), these cultural similarities most likely stem from the Circumpolar substrate shared by ancient populations in the New World, northern Asia and northern Europe and reflecting a migration across the Bering Strait, not the Atlantic.
It’s tempting to see the inflow of East Siberian lineages into Mesolithic northwest Europeans as associated with the spread of Uralic languages. As Der Sarkissian (2011, 105-106) writes,
“Even though matches were found all over Siberia, the distribution was markedly skewed towards populations of east Siberia. This was caused by the presence of the basal C* and D* haplotypes in Bolshoy Oleni Ostrov, which displayed an east-centred distribution encompassing a broad range of Eurasian populations from the Volga–Ural region, west Siberia, central Siberia to east Siberia, Mongolia, and the Altai in the South. The maximum percentage of shared haplotypes was observed with the south Siberia Tuvinians (12.2%), and interestingly the haplotypes were found to be basal (Figure 4B). The maximal shared percentage of derived haplotypes was reached for the pool of north and east Siberian Nganasans/Kets/Evenks/Yakuts (4.2%), principally due to the presence of the particular U5a1 16192T-16256T-16270T- 16399G and Z1a 16129A-16185T-16223T-16224C-16260T-16298C haplotypes. All ‘Siberian’ haplotypes as well as the ‘European’ U5a haplotype 16192T-16256T- 16270T could be found in Buryats from the peri-Baikal region (Pakendorf et al., 2006; Derenko et al., 2007). This distinct genetic link between Bolshoy Oleni Ostrov and present-day Buryats was supported by the occurrence of the rare haplotype C5 16148T-16223T-16288C-16298C-16311C-16327C in a single individual of the comparative dataset, who belongs to the Buryat population (Derenko et al., 2007).”
Uralic languages are known to share a lot of grammatical, morphological and phonological traits with Altaic languages (of which Tuvan and Buryat are part) suggesting that Uralic and Altaic, albeit not a family in and of itself, must have been in close geographic proximity with each other for a long time to enable the cross-pollination of these linguistic traits across the ethnic boundaries. But linguists tend to associate the spread of Uralic languages with the Eneolithic period.





German,
Somewhat on topic – do you care to comment on my response to Marnie’s article here???
http://linearpopulationmodel.blogspot.com/2012/12/little-big-man.html
It pertains to the Salishan-Irish connection?! 😉
Thanks, I wasn’t aware of this piece of research from DNA Tribes. It seems it goes with their global autosomal tree in which the primary split is between Amerindians and other populations (not between Africans and the rest) – see “A New Genetic Map of Interconnected World Regions” at the bottom of http://www.dnatribes.com/populations.html. I don’t know where they got their Salishan sample and how robust it is, but it does seem consistent with other studies – by academic and folk scientists – pointing to “Amerindian admixture” in (Northern) Europe (http://anthropogenesis.kinshipstudies.org/2012/09/how-europeans-got-to-be-10-american-indians/). With more ancient and modern DNA samples, with more samples from tribal populations and with increased precision in identifying variable sites, we can expect the onion of human origins to be peeled to reveal ancient connections later obscured by agropastoralist expansions.
I would consider anything of that sort found in IE-speaking populations as retentions from the earlier Mesolithic and Paleolithic populations absorbed by IE-ans in the course of their expansion.
As for Salishan-Dene-Caucasian linguistic link proposed by Shevoroshkin, it’s too hypothetical to be useful. Think about it: Na-Dene (with Haida) is poorly accepted, Dene-Caucasian is entertained by less than a handful of linguists, Salishan-Dene-Caucasian is completely a one-man show. There’s no population genetic pattern associated with Dene-Caucasian or with Salishan-Dene-Caucasian. Even in the DNA Tribes study, Basques don’t have the Salishan component, but they are part of Shevoroshkin’s Salishan-Dene-Caucasian “family.”
Genes and languages aren’t generally correlated due to language replacement processes in which a small group imposes their language to a larger population (elite dominance). But gene flow can help us trace the spread of language families associated with demic expansions. The Dene-Caucasian/Sino-Caucasian/Vasco-Caucasian macro-family/phylum (certainly not a “family”) could be one of these.
“By comparison, the modem Boreal zone cultures of Siberia seem relatively
simple (with the exception of the Yakuts). Far from resembling
these, the most convincing ethnographic parallel to Oleneostrovski society,
and indeed to the late Mesolithic period in Karelia as a whole,
might well be the complex cultures of the American Northwest Coast.”
Oleneostrovski mogilnik: Reconstructing the Social and
Economic Organization of Prehistoric Foragers in
Northern Russia
JOHN O’SHEA (1984)
http://deepblue.lib.umich.edu/bitstream/handle/2027.42/24870/0000297.pdf?sequence=1
This is a neat find Gisele! Thank you!
In the pre-print “Genomics of Mesolithic Scandinavia reveal colonization routes and high-latitude adaptation” Supplementary Figure 1 , up to 20% of some of the Mesolithic Scandinavian nuclear genomes are ‘Native American’. Sample No. EHG-UzOO74 is, in fact, the one which produced the mtDNA haplogroup C1 sequence. The nuclear DNA then bolsters the initial interpretation based upon mtDNA, as I see it.
http://www.biorxiv.org/content/biorxiv/suppl/2017/07/17/164400.DC1/164400-1.pdf
Hi Gisele, sorry for being out of touch. Which K level are you looking at in Suppl Fig 1?
I was looking at K=20 but it is visible at most K values. Yes, supplementary Figure 1 (url above).
Thanks Gisele. I can see it now clearly across the K-levels. At lower Ks you also see it in all of the ancient samples (always and high in MA-1, always and low in Mota and Ust-Ishim, sometimes and low in Satsurablia).