Ancient Kostenki 14 (Markina Gora) DNA: A Glimpse into a Population on Its Way from America to Africa
Science DOI: 10.1126/science.aaa0114
Genomic Structure in Europeans Dating Back at Least 36,200 Years
Andaine Seguin-Orlando, Thorfinn S. Korneliussen, Martin Sikora, Anna-Sapfo Malaspinas, Andrea Manica, Ida Moltke, Anders Albrechtsen, Amy Ko, Ashot Margaryan, Vyacheslav Moiseyev, Ted Goebel, Michael Westaway, David Lambert, Valeri Khartanovich, Jeffrey D. Wall, Philip R. Nigst, Robert A. Foley, Marta Mirazon Lahr, Rasmus Nielsen, Ludovic Orlando, and Eske Willerslev.
The origin of contemporary Europeans remains contentious. We obtain a genome sequence from Kostenki 14 in European Russia dating to 38,700 to 36,200 years ago, one of the oldest fossils of Anatomically Modern Humans from Europe. We find that K14 shares a close ancestry with the 24,000-year-old Mal’ta boy from central Siberia, European Mesolithic hunter-gatherers, some contemporary western Siberians, and many Europeans, but not eastern Asians. Additionally, the Kostenki 14 genome shows evidence of shared ancestry with a population basal to all Eurasians that also relates to later European Neolithic farmers. We find that Kostenki 14 contains more Neandertal DNA that is contained in longer tracts than present Europeans. Our findings reveal the timing of divergence of western Eurasians and East Asians to be more than 36,200 years ago and that European genomic structure today dates back to the Upper Paleolithic and derives from a meta-population that at times stretched from Europe to central Asia.
The Kostenki 14 (also known as Markina Gora) skeleton has yielded the oldest-to-date ancient DNA sample from West Eurasian anatomical specimens. As a 36,000 cal-year old skeleton from central Russia surrounded by generic Aurignacian and some local (Streletskyan) tools, Kostenki 14 (K14) comes from some of the most intriguing and coveted by paleontologists strata claimed by Upper Paleolithic (UP) Europeans. The skull often described by past craniologists as “unique,” “transitional,” “emergent,” “Grimaldian” and “marginal” due to its pseudo-Australoid, pseudo-Andamanese or pseudo-Negroid traits (robusticity, alveolar prognathism combined with protruding nasal bones, wide nose, small braincase, low and narrow face, deep infraglabellar notch and supraorbital tori) is clearly different from Neandertals and falls fully within UP cranial variation (Gerasimova, M. M. “Esche raz o paleoantropologicheskikh nakhodkakh v Kostenkakh.” Etnograficheskoe obozrenie 2 (2010): 26-40). I’m not aware of a study that directly compares K14 with the roughly contemporaneous Hofmeyr skull from South Africa dated at 36,000 YBP and described (Grine et al. 2007) as forming a cluster with UP Europeans to the exclusion of modern Africans (K14 was not included in Grine et al.’s UP sample). But it would be appropriate to compare the two considering that K14 shows a pronounced Sub-Saharan African-related ancestry component (see below).
It’s noteworthy that both skulls are characterized by alveolar prognathism, large nasal breadth (pyriform aperture), deep infraglabellar notch and continuous supraorbital tori – the strongest form of expression of the supraorbital ridge (see below; the Hofmeyr skull is from Grine et al. 2007). K14 is a smaller skull than Hofmeyr, though. This trait makes it look more like modern Africans, while most UP skulls are as large as Hofmeyr. Hofmeyr’s dentition is poorly preserved but it has some Sinodont traits such as large second molars (M2) (Grine et al. 2007, 227-8). K14’s teeth lack incisor shoveling (this likely means that K14 had the ancestral state of EDAR gene) and the upper second molars are large buccolingually but small mesiodistally (Khaldeeva, N. I. “Odontometricheskii analiz paleomaterialov za period Verkhnii Paleolit – Sovremennost’: V tsentre polia i po kraiam.” Etnograficheskoe obozrenie 2 (2010): 15-25).
Now, a whole-genome analysis of K14 by Seguin-Orlando et al., which came on the heels of other high-profile analyses of ancient Eurasian DNA samples such as the Malta boy (24,000 cal YBP) and Ust’-Ishim (45,000 cal YBP) has taken the analytical outlook on the biological affinities of Upper Paleolithic Europeans described above to an entirely new level. In addition to identifying Kostenki Man as a dark-skinned, dark-eyed individual, Seguin-Orlando et al.’s (2014) findings can be distilled down to the following:
1. K14’s mtDNA profile yields no surprises: it belongs to hg U2, which has patchy distribution in West Eurasia from Western Europe to India and western Siberia and has been attested in several ancient remains in Eurasia ranging from 4800 BP (Kromsdorf, Germany) to 2000 BP (Bøgebjerggård, Denmark, and the Xiongu Cemetery , northeast Mongolia). The U clade accounts for some 10% of modern European mtDNAs but was much more common in the past and is considered to be the main mtDNA signature of pre-LGM Europe whose ancient range reached into Siberia. MA-1 mtDNA at 24,000 YBP is of the U clade as well.
2. K14’s Y-DNA is thought-provoking. It belongs to hg C and is closely related to the C lineage detected in the Mesolithic La Brana sample from Spain (7,000 YBP). When first discovered, the Y-DNA C lineage in La Brana looked aberrant but now it’s clear that it constitutes an ancient West Eurasian lineage linking such distant geographies as Spain and central Russia and subsequently displaced from the European mtDNA pool by younger lineages. Hg C is much better preserved in contemporary East Asia, island Oceania, the Sahul and the New World. Importantly, hg C in K14 and La Brana is closely related to those C lineages in eastern Eurasia (hgs C1, C2 and C5 all carrying F3393/K29 mutation) that are not found in the Americas (only hg C3 with the defining M217 mutation is found there). Hg C is rare in modern Amerindians but this is likely an effect of relatively recent expansion of hg Q. Considering that hg C3* was discovered in South America and C3b is found at low frequencies across North America (peaking in Na-Dene), it’s likely that hg Q formed a wedge that minimized a once more prevalent Amerindian haplogroup. (At the same time, hg C3 found in northeast Asia could not have been a source of Amerindian C3 because much more common East Asian hgs N and O did not make it to America. This suggests that northeast Asian C3 is likely a product of a spillover from the Americas at the end of the Ice Age caused by an expansion of Y-DNA Q-bearing populations within the Americas. This means that 12,000 years ago East Eurasians only had hgs C1, C2, C4 and C5 forming a single clade opposed to America-specific hg C3.)
The story of Y-DNA lineage replacement in the New World is not too dissimilar from West Eurasia where Y-DNA hg R (a close relative of hg Q) rose to dominance only in the past 10,000 years, while hg C – once found from Spain to Russia – nearly disappeared. At 24,000 YBP hg R is attested further out east in the MA-1 of western Siberia but so far not in Europe. So, in an ironic twist of evolution, UP European Y-DNAs were similar to those of ancient Amerindians and East Eurasians (especially Oceanians and Sahulians), while Y-DNAs of modern Europeans are similar to those of modern Amerindians but not modern East Asians or Sahulians. Intriguingly, autosomal evidence, too, shows that both ancient and modern Europeans are closer to ancient and modern Amerindians than to ancient and modern East Eurasians (Raghavan et al. 2013, Fig. 3b; also Lazaridis et al. 2014), while mtDNA shows that the oldest West Eurasian clade U (attested in Kostenki as U2 and in MA-1 as U*) is closely related to hg B widely found in America and Oceania and attested in Tianyuan in southern China at 40,000 YBP but not as much in modern East Asians. Importantly, with the discovery of Y-DNA hg C in ancient Europe, the presence of various hg E lineages (E-M243, E-M96*, etc.) in modern West Eurasians yields support to an interpretation of the Y-DNA phylogeny whereby hg E-M96 colonized Africa from Eurasia. It’s also interesting that the patchy distribution of hg C1 across Eurasia, the long-distance connection between ancient West Eurasian hg C and the East Eurasian C lineages that are not C3 but the absence of hg C1 in America parallels the footprint displayed by Y-DNA hg D (the next of kin of hg E) which is found in Tibet and Japan but not much elsewhere in the Old World and never in the New World.
3. Autosomally K14 belongs with the rest of West Eurasians – ancient and modern. But this is not to be interpreted simplistically. K14 shows the strongest affinities with ancient (West) Eurasian samples such as Malta (MA-1), La Brana, Loschbour, Motala and Ajv58 across all measures. La Brana and Loschbour are especially close to K14. Among modern Europeans, f3 and D-statistic point to northern Europeans (especially, Lithuanians) as the closest living proxies for K14. However, this shared ancestry is not as strong as, for instance, the affinity between MA-1 and modern Amerindians suggesting that significant populational changes have occurred in Europe since the K14 times. In a PCA, K14 falls outside of the contemporary European genetic variation by showing an “eastward” pull and clustering with contemporary South and Central Asian populations (mtDNA hg U2, too, is presently found at elevated frequencies in South Asia). According to a clustering analysis (Fig. 1B, see on the left), K14 “plays” in all 3 autosomal admixture 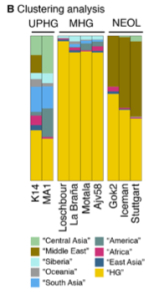 components – “West Eurasian Hunter-Gatherer” (WHG) or “Mesolithic Hunter-Gatherer” (MHG), “Ancient North Eurasian” (ANE) and “Basal European” (BEu) that Lazaridis et al. (2014) detected in contemporary and ancient West Eurasians. This has come as a surprise to many, especially to David Reich, because Lazaridis et al. (2014) and Fu et al. (2014) attributed the emergence of the BEu component across Europe to the Neolithic expansion of Middle Eastern farmers. However, K14 provides evidence that BEu has been in Europe since the Upper Paleolithic.
components – “West Eurasian Hunter-Gatherer” (WHG) or “Mesolithic Hunter-Gatherer” (MHG), “Ancient North Eurasian” (ANE) and “Basal European” (BEu) that Lazaridis et al. (2014) detected in contemporary and ancient West Eurasians. This has come as a surprise to many, especially to David Reich, because Lazaridis et al. (2014) and Fu et al. (2014) attributed the emergence of the BEu component across Europe to the Neolithic expansion of Middle Eastern farmers. However, K14 provides evidence that BEu has been in Europe since the Upper Paleolithic.
4. There is an intriguing asymmetry in the distribution of ancestry components between K14 and MA-1. Both K14 and MA-1 have South Asian and Oceanian components. K14 has an East Asian component (BLUE) that “ate” into MA-1’s stronger Central Asian (LIGHT GREEN) component. Both K14 and MA-1 stand out against later samples as having a higher diversity of high frequency ancestry components. It’s clear that over time the “Western Hunter-Gatherer” component (YELLOW) came to dominate what would become Mesolithic Europeans in post-LGM times and then its dominance got minimized by the rising prominence of the BEu component (BROWN). The combination of the two resulted in the demise of the most if not all most ancient high-frequency components such as Amerindian and Oceanian. But now, while both specimens show “Amerindian admixture” (DARKER GREEN in Fig. 1B on the left), MA-1 has a much higher concentration of it. This is consistent with the geographic placement of the two specimens: MA-1 is in South Siberia, while K14 is in central Russia. Correspondingly, K14 contains a Middle Eastern component (a.k.a. “BEu”) (BROWN) and, importantly, an African component (RED) comes along with it. So, if MA-1 skews Amerindian, K14 skews Middle Eastern and African. Both “Amerindians” and “Africans” are found in unlikely places and time periods: “Amerindians” in South Siberia at 24,000 YBP and “Africans” in the Russian Plain at 36,000 YBP. According to Lazaridis et al. (2014) and Fu et al. (2014), BEu is an ancestral population that split from non-Africans after they had left Africa but before the separation of a population represented by the most ancient Eurasian sample, namely Ust’-Ishim (45,000 YBP from western Siberia). BEu was supposed to have stayed in the Middle East, in close proximity to Africa, only to expand into greater West Eurasia with the advent of farming. But now it has popped chronologically some 25,000 years earlier than expected, geographically in much more northern latitudes and genetically in conjunction with what looked to out-of-Africanists as the younger “Ancient North Eurasian” (ANE) and “Western Hunter-Gatherer” (WHG) components.
Seguin-Orlando et al. (2014) make a critical observation to the effect that all non-Africans are more like MA-1 than K14:
“We find that all contemporary non-Africans, except Australo-Melanesians, are closer to either Mal’ta (MA1) or MHGs than to K14 [e.g., Z = -5.3, for D (Mbuti, Han; Loschbour, K14); SOM S9; table S10; fig. S19].”
They refer to the following plots from S19. All the populations are shifted left (toward MA-1 or Loschbour) to a varying degree.
They also observe that East Asians are equidistant from both UP Europeans and Neolithic Europeans.
“Consistent with this hypothesis, we find that East Asians are equally distant to NEOL and K14 using D-statistics as described above [e.g., Z = 0.0, for D (Mbuti, Han; Stuttgart, K14); tables S10 and S11].”
This can be seen in the last plot from S19. East Asians (Dai and Han) are at 0 in a plot anchored in Stuttgart and Kostenki.
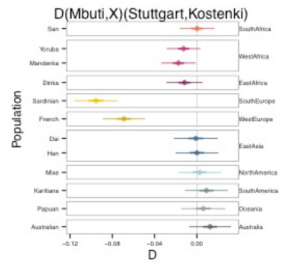 This leads Seguin-Orlando et al.’s (2014) to their most dramatic conclusion, namely that K14 is more basal than the non-African clade and this is due to the presence of the BEu component in K14. But what they have overlooked is the position of Sub-Saharan Africans on the same plots! San, Yoruba, Mandenka and Dinka are all shifted toward MA-1 over K14. So, contrary to Seguin-Orlando et al.’s (2014), MA-1 is “preferred” not just by most non-Africans but also by all Sub-Saharan Africans. Africans are also more Neolithic-European shifted than Kostenki-shifted but this applies more to Yoruba, Mandenka and Dinka and less so to San. San, just like East Asians, are equidistant from Neolithic Europeans and Paleolithic Europeans. This means that, when Africans are factored in, MA-1, with its pronounced “special affinity” to Amerindians and no connection to East Asians, becomes basal to all modern humans, while K14 becomes an ancient representative of a clade that comprises two subclades: 1) Upper Paleolithic Europeans, Neolithic Europeans/modern Middle Easterners, East African pastoralists (Dinka) and West African farmers (Yoruba, Mandenka); 2) East Asians and South African hunter-gatherers (San). Fig. S19c also shows that Australians, Oceanians and Amerindians are absolutely devoid of the BEu component (also present in K14) that’s prominent in modern Western Europeans but also in West Africa and East Africa. But this BEu component could not have come from the Middle East because modern Lithuanians are closer to K-14 than Stuttgart (Fig. S14), the alleged recipient of the Middle Eastern gene flow. So, the Middle Eastern populations must have received it from an UP European population related, first, to K-14, then to MA-1 and, finally, to Amerindians. This is why, while MA-1 ancestry is found across Sub-Saharan Africa, the BEu part of K14 ancestry is absolutely missing among Amerindians and Sahulians. This proves that Africa was colonized from Eurasia by modern humans, while Amerindians and Sahulians are the two most unadmixed and isolated populations among modern humans. The importance of K14 for modern human origins, therefore, is that it represents a pivotal population in a state of transition from a New World ancestor of all modern humans to an ancestor of all modern Sub-Saharan African populations.
This leads Seguin-Orlando et al.’s (2014) to their most dramatic conclusion, namely that K14 is more basal than the non-African clade and this is due to the presence of the BEu component in K14. But what they have overlooked is the position of Sub-Saharan Africans on the same plots! San, Yoruba, Mandenka and Dinka are all shifted toward MA-1 over K14. So, contrary to Seguin-Orlando et al.’s (2014), MA-1 is “preferred” not just by most non-Africans but also by all Sub-Saharan Africans. Africans are also more Neolithic-European shifted than Kostenki-shifted but this applies more to Yoruba, Mandenka and Dinka and less so to San. San, just like East Asians, are equidistant from Neolithic Europeans and Paleolithic Europeans. This means that, when Africans are factored in, MA-1, with its pronounced “special affinity” to Amerindians and no connection to East Asians, becomes basal to all modern humans, while K14 becomes an ancient representative of a clade that comprises two subclades: 1) Upper Paleolithic Europeans, Neolithic Europeans/modern Middle Easterners, East African pastoralists (Dinka) and West African farmers (Yoruba, Mandenka); 2) East Asians and South African hunter-gatherers (San). Fig. S19c also shows that Australians, Oceanians and Amerindians are absolutely devoid of the BEu component (also present in K14) that’s prominent in modern Western Europeans but also in West Africa and East Africa. But this BEu component could not have come from the Middle East because modern Lithuanians are closer to K-14 than Stuttgart (Fig. S14), the alleged recipient of the Middle Eastern gene flow. So, the Middle Eastern populations must have received it from an UP European population related, first, to K-14, then to MA-1 and, finally, to Amerindians. This is why, while MA-1 ancestry is found across Sub-Saharan Africa, the BEu part of K14 ancestry is absolutely missing among Amerindians and Sahulians. This proves that Africa was colonized from Eurasia by modern humans, while Amerindians and Sahulians are the two most unadmixed and isolated populations among modern humans. The importance of K14 for modern human origins, therefore, is that it represents a pivotal population in a state of transition from a New World ancestor of all modern humans to an ancestor of all modern Sub-Saharan African populations.
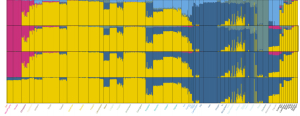
This interpretation is supported by the ADMIXTURE plot (Fig. S20, K=2 through K=5 are reproduced below next to a plot from Rosenberg et al. 2002):
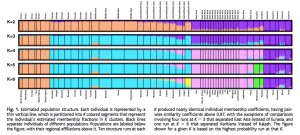 Amerindians are at all levels an “unadmixed” population. At K=2 their DARK BLUE color is present in Sub-Saharan Africa, including San. Among ancient samples MA-1 shows increased concentration of this Amerindian component. At the same time, the YELLOW component (high in Africa, Middle East and among European Neolithic populations) is altogether missing from the New World suggesting that it evolved after modern humans had left America.
Amerindians are at all levels an “unadmixed” population. At K=2 their DARK BLUE color is present in Sub-Saharan Africa, including San. Among ancient samples MA-1 shows increased concentration of this Amerindian component. At the same time, the YELLOW component (high in Africa, Middle East and among European Neolithic populations) is altogether missing from the New World suggesting that it evolved after modern humans had left America.
5. Seguin-Orlando et al. (2014) discovered a greater proportion of Neandertal alleles in K14 and its closest ancient “cousin,” the La Brana skull, compared to other ancient and modern populations (see below). This jibes well with the recent report that another ancient Eurasian sample, Ust’-Ishim at 45,000 YBP, also has a greater affinity to Neandertals than living humans.
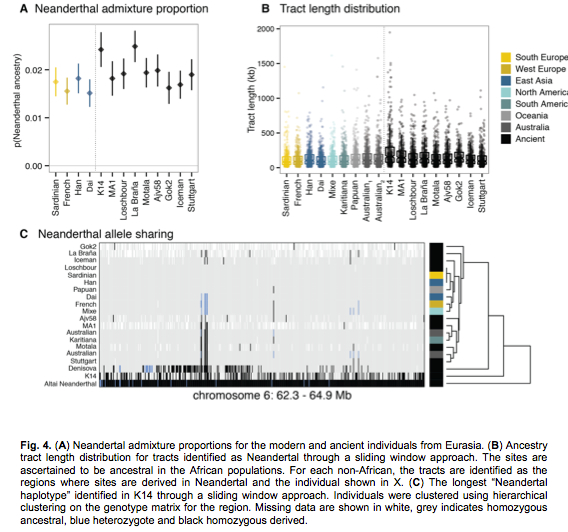 However, on Fig. 4A, Amerindians are strangely missing. On Fig. 4B K14 stands out as having the longest chunks of DNA in common with Neandertals. It’s followed by a group of populations that includes Karitiana, Mixe, Australian and Papuans, all showing the longest Neandertal-like chunks among modern populations. Fig. 4C gives a different view of Neandertal allele sharing on chromosome 6, with K14 clearly maintaining a stronger continuity with Neandertals. It’s remarkable, however, that one region of chromsome 6 shows pronounced Neandertal continuities also in Stuttgart, Australian, Motala and Karitiana sequences (derived homozygotes) and in Dai, French and Mixe sequences (heterozygotes). It’s not clear, however, why on the resulting tree these populations with pronounced Neandertal affinities ended up more downstream from the root of the tree than those populations that do not show such a pronounced continuity.
However, on Fig. 4A, Amerindians are strangely missing. On Fig. 4B K14 stands out as having the longest chunks of DNA in common with Neandertals. It’s followed by a group of populations that includes Karitiana, Mixe, Australian and Papuans, all showing the longest Neandertal-like chunks among modern populations. Fig. 4C gives a different view of Neandertal allele sharing on chromosome 6, with K14 clearly maintaining a stronger continuity with Neandertals. It’s remarkable, however, that one region of chromsome 6 shows pronounced Neandertal continuities also in Stuttgart, Australian, Motala and Karitiana sequences (derived homozygotes) and in Dai, French and Mixe sequences (heterozygotes). It’s not clear, however, why on the resulting tree these populations with pronounced Neandertal affinities ended up more downstream from the root of the tree than those populations that do not show such a pronounced continuity.




I just wanted to add that the frequencies of mtDNA haplogroup U2 in India and surrounding areas have been illustrated in the following recent article:
Indian Ocean Crossroads: Human Genetic Origin and
Population Structure in the Maldives
Pijpe et al, (2013) AJPA
http://onlinelibrary.wiley.com/doi/10.1002/ajpa.22256/abstract
See Figure 2 and also the supplementary files.
Nice, thanks Gisele. I found the article you mentioned.
”From the Y-DNA perspective, the earliest West Eurasians were apparently similar to East Eurasians and Amerindians and their distinctive West Eurasian genetic and phenotypic makeup is a product of post-LGM” How is this logical? You see if ancient European C1a-(V20) is 55,000 years away from American C3 (C2), than European R is around 32,500 years away from Native american Q. On top of that like 99% of Native Americans have Q with only like 1% having C3(C2). You should say this “”From the Y-DNA perspective, the earliest West Eurasians were apparently similar to Melanesians however in the last 6000 years they have come to be very similar (on the male side) to Native Americans, while still retaining some similarity to Melanesians”
Why does every person always deny how close Q and R are? It is really agitating.
Thanks, good thinking. I updated my interpretation of Y-DNA evidence per your feedback. Let me know how it looks now. But please follow this blog’s policy and post with real credentials (if not academic, then at least personal ones).
I think the updates are great.
Wait… “Into Africa”… Sub-Saharan even? And from America? And without carrying the genetic legacy of Neanderthal or Denisovan?
Please clarify… seems to fly in the face of all evidence… burden of proof and all…
The frequencies of Neandertal and Denisovan alleles (everywhere low) decline from East to West (PNG, America, East Asia) > Europe > Africa. The fact that alelles at small frequencies decline and get altogether lost is a pattern. Why would this be surprising?