
The Best Kept Secret in Populaton Genetics, or Truth about African Genetic Diversity
Nature (2014) doi:10.1038/nature13997
The African Genome Variation Project Shapes Medical Genetics in Africa
Gurdasani, Deepti, Tommy Carstensen, Fasil Tekola-Ayele, Luca Pagani, Ioanna Tachmazidou, Konstantinos Hatzikotoula, Savita Karthikeyan, Louise Iles, Martin O. Pollard, Ananyo Choudhury, Graham R. S. Ritchie, Yali Xue, Jennifer Asimit, Rebecca N. Nsubuga, Elizabeth H. Young, Cristina Pomilla, Katja Kivinen, Kirk Rockett, Anatoli Kamali, Ayo P. Doumatey, Gershim Asiki, Janet Seeley, Fatoumatta Sisay-Joof, Muminatou Jallow, Stephen Tollman, Ephrem Mekonnen, Rosemary Ekong, Tamiru Oljira, Neil Bradman, Kalifa Bojang, Michele Ramsay, Adebowale Adeyemo, Endashaw Bekele, Ayesha Motala, Shane A. Norris, Fraser Pirie, Pontiano Kaleebu, Dominic Kwiatkowski, Chris Tyler-Smith, Charles Rotimi, Eleftheria Zeggini, and Manjinder S. Sandhu.
Given the importance of Africa to studies of human origins and disease susceptibility, detailed characterization of African genetic diversity is needed. The African Genome Variation Project provides a resource with which to design, implement and interpret genomic studies in sub-Saharan Africa and worldwide. The African Genome Variation Project represents dense genotypes from 1,481 individuals and whole-genome sequences from 320 individuals across sub-Saharan Africa. Using this resource, we find novel evidence of complex, regionally distinct hunter-gatherer and Eurasian admixture across sub-Saharan Africa. We identify new loci under selection, including loci related to malaria susceptibility and hypertension. We show that modern imputation panels (sets of reference genotypes from which unobserved or missing genotypes in study sets can be inferred) can identify association signals at highly differentiated loci across populations in sub-Saharan Africa. Using whole-genome sequencing, we demonstrate further improvements in imputation accuracy, strengthening the case for large-scale sequencing efforts of diverse African haplotypes. Finally, we present an efficient genotype array design capturing common genetic variation in Africa.
Gurdasani et al. (2014) is a very important paper as it dispels with one sway four long-standing and hard-working myths, namely that 1) Africa is the most genetically diverse continent; 2) genetic diversity is an indicator of population age; 3) non-African diversity is a subset of African diversity; 4) serial bottlenecks out of Africa are responsible for the observed global patterns of genetic diversity.
Fundamentally, there are two kinds of genetic diversity: intergroup (between-group, among-groups) diversity and intragroup (within-group) diversity. The two diversity measures are dialectically intertwined, so that an increase in one kind of diversity leads to a decrease in the other kind of diversity. As divergent populations merge, they lose some of their intergroup diversity and become more similar to each other but they gain intragroup diversity because now they are enriched with two or more sets of alleles that evolved separately during the time the populations were isolated from each other. As populations drift apart, their intergroup diversity increases, while their intragroup diversity decreases as alleles get lost through drift.
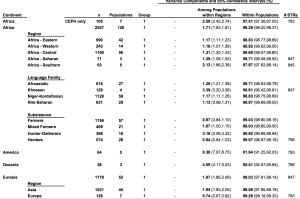
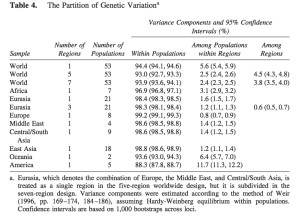
It has become a truism (pace Lewontin) that most genetic variation (some 85%) among humans happens within populations and not between them. The 15% of variation that happens between populations is claimed to be indicative of the young age of the human species. So, it’s intergroup diversity that’s associated with population age and not intragroup diversity (see more here). Continental populations are uneven in their apportionment of inter- vs. intragroup diversity. Sub-Saharan Africans are rich in only one kind of diversity – intragroup diversity, which is a function of effective population size and may reflect (as in the case of admixed post-1492 New World populations that are also rich in intragroup diversity or heterozygosity) layers of admixture. When it comes to intergroup diversity, Africans are only moderately differentiated and so are far from being the “most diverse” among human populations. Amerindians who are the most distant from Africa geographically are the exact opposite from Africans genetically: they have the lowest intragroup diversity among all human continental groups but world highest intergroup diversity values. This pattern is fully captured in the following tables from Tishkoff et al. 2009 (left) and Rosenberg, Noah. “A Population-Genetic Perspective on the Similarities and Differences Among Worldwide Human Populations,” Human Biology 83, no. 6 (2011), 670 (right).
Gurdasani et al. (2014) drill deeper into this remarkable pattern. They provide a composite table (Suppl Table 1) of worldwide Fst (fixation index) values. (The Africa-America Fst is unusually low, which suggest that they used an Amerindian sample that’s admixed with African blacks. Fst between unadmixed Amerindians and Africans reaches 0.281; see Verdu et al. 2014 “Patterns of Admixture and Population Structure in Native Populations of Northwest North America.” PLoS Genet 10(8): e1004530). The limited extent of intergroup differentiation in Africa is evident if we compare Fst across all Khoisan groups (0.019) or between Nilo-Saharan- and Khoisan-speakers (0.058) with Fst between two Amerindian tribes such as Pima and Seri (0.094) (Verdu et al. 2014).
Recently expanded populations within Africa, such as Niger-Congo and Afroasiatic speakers, have one of the lowest intergroup diversity values within Africa. Foragers such as Khoisan have higher intergroup diversity values and are therefore more Amerindian-like than farmers. Niger-Congo and Afroasiatic populations show intragroup diversity that’s higher than that of Khoisan (see Tishkoff et al.’s table above). It’s the same pattern as I observed between Taiwanese aborigines and Polynesians: the former are more homozygous than the latter, hence the colonization of Polynesia from Taiwan was accompanied by heterozygosity increase, not heterozygosity reduction, as a serial bottleneck approach to interpreting worldwide patterns of genetic diversity would suggest.
Gurdasani et al. (2014) ran several statistics (straight f4, f4 with ancestry masking, Z-score and ADMIXTURE) to test for Eurasian gene flow in Sub-Saharan Africa and found that it reaches 50% in some populations. The oldest trace of Eurasian admixture in Africa (7,500-10,000 years ago) was found in Yoruba, which is consistent with previous finding of Neandertal genes in Yoruba. East Africa is the region most affected by Eurasian admixture. Among all the statistics, ADMIXTURE runs proved to yield the most evidence for Eurasian gene flow in Sub-Saharan Africa, which suggests that the other statistics may be prone to genetic drift that can change negative values to positive and hence obfuscate ancient admixture. Eurasian admixture increases African Fst and, once it’s masked, mean pairwise Fst in Africa declines from 0.021 to 0.015, which is similar to Fst values encountered between geographically separated populations in Europe. They identified two “pockets” of elevated Fst (after masking for Eurasian ancestry) – one in Ethiopia and the other one in West Africa. In the latter case, West African Igbo showed admixture with a foraging population, which is closer to Khoisan than to Pygmies.
Finally, another gem into my Kunstkamera collection. Writing about Gurdasani et al. (2014), Razib “Cat Lady” Khan made a ridiculous claim:
“the genetic variation across African populations once you remove Eurasian ancestry is not that high. This is curious in light of the truism that most genetic variation in humans is found within Africa, but as Nick Patterson pointed out to me years ago: this applies to variation within populations, not across them.”
Nick Patterson must be the chief secret keeper among population geneticists because he never published anything to this effect. Instead, he chose to whisper this secret into the ear of no one else but Razib Khan. The truth is that it’s me and not Nick who mentored Razib on this matter and did it in a public forum. This happened in the Comments section of Gene Expression and, according to the log, it was 4 years ago.
 It used to be that Razib’s knowledge of genetics was poor. Now that it has improved it’s his memory that’s failing him.
It used to be that Razib’s knowledge of genetics was poor. Now that it has improved it’s his memory that’s failing him.



Good one. “Cat Lady”. Classic.
Can you tell me if studies have been done with Asians and other groups comparable to this one?
The bonobo genome compared with the chimpanzee and human genomes
http://www.nature.com/articles/nature11128
I’m not sure how you can claim its a myth that Africa is the most genetically diverse continent in the world when the study you linked stated:
“Although Africa is the most genetically diverse region in the world, we provide evidence for relatively modest differentiation among populations representing the major sub-populations in SSA, consistent with recent population movement and expansion across the region beginning around 5,000 years ago—the Bantu expansion.”
And your other claim needs to be addressed: Native American populations possess more genetic differentiation between each than Tropical African populations. Would this not simply be even more indicative of population bottlenecks in Native Americans? Not to mention that they lack intragroup genetic diversity. Is this not the exact result of population bottlenecks? Genetic differentiation from the original population, but a decreased overall genetic diversity?
From the study you cited, it simply seems more likely that the Bantu Expansion and the lack of intergroup diversity is more reflective of a complex series of migrations originating from West Africa into the rest of Africa rather than an abrupt genetic bottleneck that resulted in specifically genetically differentiable populations.
Thanks for your comments. Per the blog’s posting rules, please sign in using your real name in the future. You bring up a critical point in the business of interpreting the global patterns of genetic diversity. SSAfricans are most genetically diverse when it comes to intragroup diversity (the fact overblown in population genetics research as a sure sign of population antiquity), but Amerindians are most diverse when it comes to intergroup diversity (the fact downplayed by population genetics research and, even when brought up, dismissed as a sign of a recent bottleneck). But now that ancient DNA is becoming increasingly available we’ve discovered that Neandertals and Denisovans, who are “older” than SSAfricans, show low intragroup diversity and high intergroup diversity. So, the myth of African antiquity as indicated by their pattern of genetic diversity has been busted by ancient DNA data from Eurasian hominins. This leaves another opportunity on the table to help us explain high intragroup diversity in SSAfricans – Eurasians who colonized Africa may have absorbed an archaic African substratum that was more diverse than Eurasian hominins. So, although some of African lineages are indeed ancient, they ended up in the modern human pool through a relatively recent admixture that would have happened when African was colonized from Eurasia. The “Amerindian bottleneck”, on the other hand, is not recent but in fact a “continuation” of the Eurasian hominin bottleneck. Amerindians have simply retained low effective population size since Mid-Pleistocene times.
Well, “Asim” is my middle name, so I guess I’ll just go by Asim.
I see your point about Neanderthal and Denisovan genetic diversity, but once again, that is simply indicative of repeated genetic bottlenecks; you must keep in mind that the ancestors of Neanderthals and Denisovans also originated in Africa, but the ancestors of modern humans remained in Africa. Technically, “Sub-Saharan Africans” are as old as Homo Sapien Sapiens themselves; there’s no evidence that aside from the common ancestral original of all humans, that Tropical Africans have undergone significant genetic unification, similar to Eurasians. However, the reason Tropical Africans are ultimately more diverse than Eurasians has nothing (directly) to do with age, but more to do with a lack of genetic bottleneck. As humans migrate, especially during the Paleolithic, they tend to only take part of the genetic diversity that the original population had to offer (because the entire population itself does not migrate, only a few select individuals from the population migrate). While this event makes the new population genetically distinguished from the original population, the overall genetic diversity of the new population is less than the original. Therefore, this explains the reason that Native Americans show the least intragroup diversity, but the most intergroup diversity. As Native Americans migrated further South the American continents, they perpetually created genetically distinct populations, with ultimately less intragroup diversity due to the bottleneck effect. This is evident due to the fact that intragroup diversity diminishes as one goes South the American continents.
This also explains how Neanderthals and Denisovans also have less intragroup diversity, but more intergroup diversity (which simply means multiple genetically distinct populations). Neanderthals and Denisovans migrated throughout Eurasia (originating from Africa), which created many genetically distinct populations as they migrated, but a diminished overall genetic diversity. Meanwhile, the ancestors of modern humans remained in Africa and did not migrate (causing genetic bottlenecks), hence had more genetic diversity than Neanderthals and Denisovans.
Theoretically, I don’t think we have any disagreements. Practically, however, the mainstream model of a serial bottleneck out of Africa that you’re advocating for is increasingly unpopular. Ancient DNA showed that Europe has experienced an increase in intragroup diversity (heterozygosity) – not a decrease as your model predicted – through the mixture of several ancestral components. Also, modern history shows that when Africans were brought to the New World as “slaves” in post-1492 times, although only West Africa got sampled, genetic diversity of the resulting African American population did not decrease but stayed relatively stable and hence representative of the overall SSAfrican diversity. This is why the earliest mtDNA studies that used African Americans as representatives of original SAAfricans generated the same tree typologies as the later studies that used SSAfricans as a sample. Even such ancient Y-DNA lineages as A00 were discovered in both West Africa and among African Americans. At the same time, there are no deep-rooting African lineages found in any populations outside of Africa along the putative migration routes out of Africa 100-50,000 years ago. Neither have they been found in ancient remains outside of Africa. E.g., Oase mtDNA is not L0, L1, L2, L4, L5 or L6 – it’s somewhere between M and N. Under my scenario of genetic admixture with more populous African archaics, it becomes clear how 1) admixture drives intragroup diversity up; and why 2) no deep-rooting African lineages have been found outside of Africa. I don’t deny that Neandertals and Denisovans have an ultimate African origin, but with the interdisciplinary data that we have for modern humans (linguistic diversity, which is a marker of behavioral modernity, is world-highest in America and Papua New Guinea and that’s precisely where we find the highest intergroup genetic diversities) an onset of modern humanity in the New World (or in a broader Circumpacific zone) and a migration into Africa seems to be more logical. (BTW, low population size – hence likely high genetic intergroup and low genetic intragroup diversity – east of the Movius linen compared to Europe and Africa has been documented by archaeologists since Lower Plaeolithic.)
Essentially, the study you cited is stating, “There is no distinct genetic difference that can be pinpointed between certain African populations like the Yoruba and the Zulu. Any genetic differentiation that is notable is typically due to admixture from Eurasians are isolated Hunter-Gatherer groups.” This is because, unlike Native Americans, Niger-Congo populations were not subject to population bottlenecks during the formation of their people groups. Because the Bantu Expansion resulted in countless complex migrations, rather than abrupt bottlenecks, no genetic differentiation from the original population was created, which would ultimately create a reduction in genetic diversity. This similar effect seems to be present in Afro-Asiatic speakers, who mostly migrated across Africa throughout the Neolithic Revolution. However, because of this, the resulting populations tend to maintain the same genetic diversity of the original.