The Origin of mtDNA haplogroup B: 9-bp deletion in America, Asia and Africa
PLoS ONE 7(2) 2012: e32179. doi:10.1371/journal.pone.0032179
Complete Mitochondrial DNA Analysis of Eastern Eurasian Haplogroups Rarely Found in Populations of Northern Asia and Eastern Europe
Derenko M., Malyarchuk B., Denisova G., Perkova M., Rogalla U., et al.
Abstract. With the aim of uncovering all of the most basal variation in the northern Asian mitochondrial DNA (mtDNA) haplogroups, we have analyzed mtDNA control region and coding region sequence variation in 98 Altaian Kazakhs from southern Siberia and 149 Barghuts from Inner Mongolia, China. Both populations exhibit the prevalence of eastern Eurasian lineages accounting for 91.9% in Barghuts and 60.2% in Altaian Kazakhs. The strong affinity of Altaian Kazakhs and populations of northern and central Asia has been revealed, reflecting both influences of central Asian inhabitants and essential genetic interaction with the Altai region indigenous populations. Statistical analyses data demonstrate a close positioning of all Mongolic-speaking populations (Mongolians, Buryats, Khamnigans, Kalmyks as well as Barghuts studied here) and Turkic-speaking Sojots, thus suggesting their origin from a common maternal ancestral gene pool. In order to achieve a thorough coverage of DNA lineages revealed in the northern Asian matrilineal gene pool, we have completely sequenced the mtDNA of 55 samples representing haplogroups R11b, B4, B5, F2, M9, M10, M11, M13, N9a and R9c1, which were pinpointed from a massive collection (over 5000 individuals) of northern and eastern Asian, as well as European control region mtDNA sequences. Applying the newly updated mtDNA tree to the previously reported northern Asian and eastern Asian mtDNA data sets has resolved the status of the poorly classified mtDNA types and allowed us to obtain the coalescence age estimates of the nodes of interest using different calibrated rates. Our findings confirm our previous conclusion that northern Asian maternal gene pool consists of predominantly post-LGM components of eastern Asian ancestry, though some genetic lineages may have a pre-LGM/LGM origin.
Link (Free Full Text PDF)
A team of Russian and Polish geneticists revisited the phylogeography of North, East and Southeast Asian mtDNA haplogroups through the lens of a massive 5000-individual-strong sample. Apart from the detected Holocene-age gene flow between Siberia and Eastern Europe, the paper takes an in-depth look into the phylogeny of haplogroup B in North Asia (see below).
Derenko et al. support the “Beringian incubator” model of the peopling of the New World: a small founding population was isolated in Beringia until about 17,000 years ago when it migrated southward and colonized the Americas. Hg B has always caused problems for any model of the peopling of the Americas because of its scarcity of in North Asia and northern North America and its high frequency in the southern areas of the New World. Derenko et al. confirm that the origin of the American Indian-specific hg B2 remains a mystery:
“It should be noted that we have not found in northern Asia any haplogroup B mtDNA lineages ancestral to Amerindian-specific B2 branch. The only Tubalar mtDNA described previously by Starikovskaya et al., designated there as B1 and interpreted as “closely related to Amerindian-specific B2 branch”, belongs in fact to the northern Asian-specific subcluster B4b1a3 which in turns is a part of major subcluster B4b1, distributed predominantly in eastern Asia. Thus, there is no evidence at this time for the occurrence of haplogroup B2 mtDNA ancestors in Siberia…”
While Derenko et al. managed to date the North Asian B4b1a3 node at ~18–20 kya using different mutation rates and ascertain a pre-LGM origin of this subcluster, they didn’t find the appropriate ancestor of American Indian hg B2, which should be expected to be of similar age to B4b1a3 if their model of the peopling of the Americas is true. American Indian hg B2, therefore, joins another American Indian mtDNA haplogroup, namely X2, in not having antecedents in Siberia.
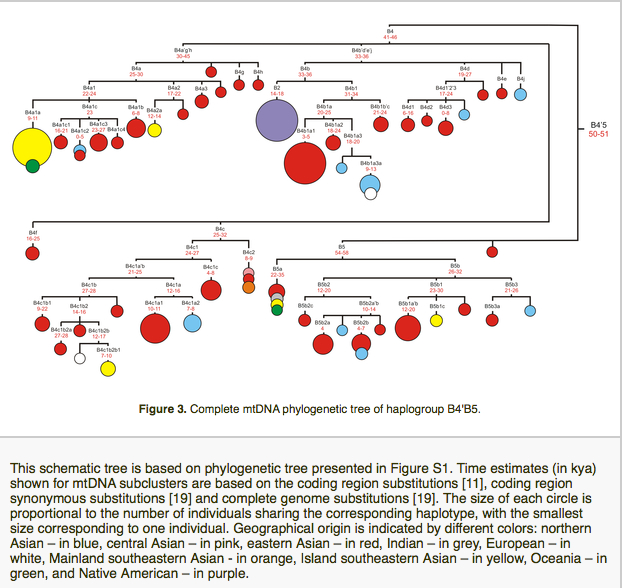
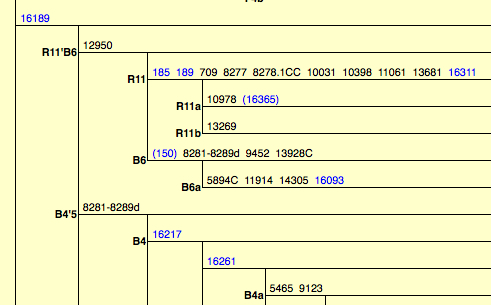
The paper also leads to an intriguing observation regarding the phylogeny of hg B. Let’s take a look at the way Derenko et al. define haplogroup B:
“Haplogroup B is identified by the presence of a 9-bp deletion in the COII/tRNALys intergenic region of mtDNA. Despite the [fact that. – G.D.] 9-bp deletion has a high recurrence, it seems that together with transition 16189 it defines fairly well a monophyletic cluster, which consists of two subhaplogroups, B4 and B5. A sister clade of B4’B5, keeping the 16189 mutation and having additional polymorphism at np 12950, has been detected in eastern and Island southeastern Asia, being designated as R11’B6. R11’B6 cluster is further subdivided in R11, lacking the 9-bp deletion, and B6, having this deletion. It is worthwhile to mention that R11 mtDNAs have been detected mainly in China, whereas B6 lineages are present both in eastern and Island southeastern Asia.”
A snapshot of this phylogenetic node can be obtained from PhyloTree:
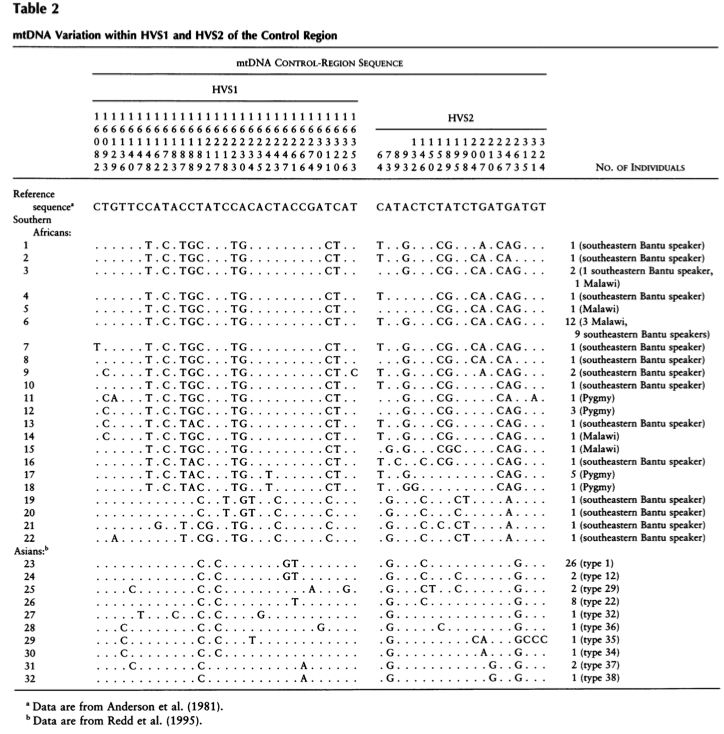
It’s clear that hg B4’B5 is a monophyletic clade formed by 9-bp deletion between np 8281 and 8289 and a C at np 16189. Interestingly, the 9bp deletion seems to recur within the sister clade R11’B6, which means that the unique mutation occurred twice on two closely related lineages, B4’B5 and B’6. In and of itself, this sounds highly suspect and suggests the mutational order may need to be revised. Extra complexity comes with the observation that the combination of 1) 9-bp deletion in the COII/tRNALys intergenic region of mtDNA between np 8281 and 8289 and 2) 16189C occurs in Sub-Saharan Africa among Pygmies and Bantu. The current mtDNA nomenclature classifies this other 9-bp deletion lineage as L0a2. Here’s a snapshot of Asian and African 9-bp deletion carrying lineages from Soodyall et al. “mtDNA Control-Region Sequence Variation Suggests Multiple Independent Origins of an “Asian-Specific” 9-bp Deletion in Sub-Saharan Africans,” Am. J. Hum. Genet. 58 (1996): 595-608.
Soodyall et al. correctly observe (p. 603) that such typical Southeast Asian and Polynesian substitutions as 16217 (T>C), 16247 (A>G) and 16261 (C>T) are not found on Sub-Saharan African 9-bp deletion-carrying lineages. But as their Table 2 (see above) clearly shows, C at np 16189 is fixed on both Asian and African sequences. And, according to Derenko et al and PhyloTree, 9-bp deletion and 16189C are the only two mutations that define hg B in Asia and America. We can see that the African sequences are very divergent and generally don’t match the Asian sequences, which led Soodyall et al. to argue for multiple origins of 9-bp deletion in world populations, but with better screening in Asia we now know that even in Asia the divergence is great and, moreover, the combination 8281-8289d + 16189C is shared between Asian B4’B5 and African L0a2 but not between B4’B5 and R11’B6 within Asia. This means that African L0a2 can be thought of as a sister of Asian hg B4’B5, and 8281-8289d and 16189C will be the defining mutations of this clade. Notably, np 16189 is hypervariable in humans but it’s fixed as C in Neanderthals (see Hagelberg E. “Recombination or Mutation Rate Heterogeneity: Implications for Mitochondrial Eve,” Trends in Genetics 2003 19 (2):84-90). mtDNA hg X is also 16189C. One may argue back that the isolation of 8281-8289d and 16189C into a single clade overlooks the “upstream” mutations leading to, respectively, B4’B5 and L0a2, but, as the example of 8281-8289d in B6 suggests, 8281-8289d may need to be re-considered as a more upstream mutation. 16223, one of the defining mutations of macrohaplogroup R, is a recurrent mutation in Africa. Sequences 19-20 in Soodyall et al.’s Table 2 carry the same mutation at 16223 (C) as all of the 9-bp deletion sequences in Asia.
The combination 8281-8289d + 16189C also occurs on the West Asian hg T2f (Pike et al. “mtDNA Haplogroup T Phylogeny based on Full Mitochondrial Sequence,” Journal of Genetic Genealogy, 6 (1), 2010, p. 9). T2f is part of hg T found in Mesopotamia, Syria, Turkey and among Bulgarian Jews (see Behar et al. “Counting the Founders: The Matrilineal Genetic Ancestry of the Jewish Diaspora,” PLoS ONE 3 (4), 2008). Its provenance, therefore, is mid-way between coastal East Asia/New World (B4’B5) and Sub-Saharan Africa (L0a2).
Behar et al. (“The Genographic Project Public Participation Mitochondrial DNA Database,” PLoS Genetics 3 (6), 2007) collected instances of 8281-8289d occurring across all existing haplogroups (see below, from Supplementary Material).
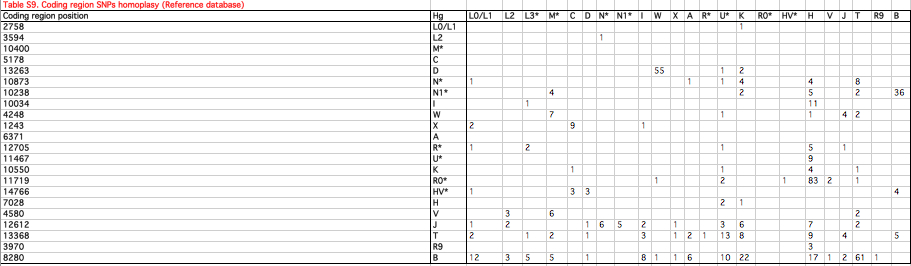 Behar et al. were looking to make a judgment as to whether some ostensibly recurrent mutations are products of recombination (whereby one lineage “donates” its portion to another lineage) or homoplasy. Since apparently 9-bp deletion is nowhere accompanied by other inconsistencies in the phylogenetic branch, Behar et al. concluded that 8281-8289d mutations are homoplastic (p. 1089). But they never considered the possibility that 9-bp deletion is a relic state that preceded the formation of all the lineages it’s currently found on and that its absence on other lineages is a product of a homoplastic 9-bp insertion. This wouldn’t create further inconsistencies in the branches, but it would redefine the branches. If in Asia and America just two states (8281-8289d and 16189C) are sufficient to define the B4’B5 clade and precisely these two states are found also in West Asia and Sub-Saharan Africa, then why not consider the possibility that the combination in question is a relic pattern? Then some regions/haplorgoups would show 9-bp insertion but keep the original 16189C, others would do the opposite. It’s hard to say for sure without re-hashing the whole mtDNA phylogeny, but if genetic labs test only some alternatives but not others it’s impossible to be certain their conclusions are right.
Behar et al. were looking to make a judgment as to whether some ostensibly recurrent mutations are products of recombination (whereby one lineage “donates” its portion to another lineage) or homoplasy. Since apparently 9-bp deletion is nowhere accompanied by other inconsistencies in the phylogenetic branch, Behar et al. concluded that 8281-8289d mutations are homoplastic (p. 1089). But they never considered the possibility that 9-bp deletion is a relic state that preceded the formation of all the lineages it’s currently found on and that its absence on other lineages is a product of a homoplastic 9-bp insertion. This wouldn’t create further inconsistencies in the branches, but it would redefine the branches. If in Asia and America just two states (8281-8289d and 16189C) are sufficient to define the B4’B5 clade and precisely these two states are found also in West Asia and Sub-Saharan Africa, then why not consider the possibility that the combination in question is a relic pattern? Then some regions/haplorgoups would show 9-bp insertion but keep the original 16189C, others would do the opposite. It’s hard to say for sure without re-hashing the whole mtDNA phylogeny, but if genetic labs test only some alternatives but not others it’s impossible to be certain their conclusions are right.
If mtDNA B4’B5 and L0a2 are indeed related, it will establish a clear mtDNA signature tying Sub-Saharan Africans, including Pygmies, with Southeast Asia, Oceania and the western parts of the New World where hg B2 predominates (it reaches fixation in several Andean populations). Ethnologists have been searching for a link between Sub-Saharan Africa, Oceania and South America for a long-time. From the musical perspective, Victor Grauer sees a South American Indian vocal style called “canonic-echoic” as a “development” of Pygmy-Bushmen vocalizing with clear parallels in Melanesia and Papua New Guinea. The same applies to pan-pipe ensembles:
“In almost all cases, from Africa to Melanesia to the Americas, the music played by such instruments is divided into at least two hocketing/interlocking parts. In the case of pipes, whistles, trumpets and horns, each instrument is capable of producing only one or two notes, but even in panpipe or flute ensembles all instruments perform in closely interactive hocketed interlock. Unlike Western counterpoint, but very similar to certain practices found in Africa, what we hear is not the intertwining of independent lines, but a resultant melodic/ polyphonic texture produced by the juxtaposition of interlocking parts. In almost all cases, from both Melanesia and the Americas, the instruments are symbolically divided into two complementary groups, one male, the other female.”
Not surprisingly, just like mtDNA hg B (and hg X) is virtually missing from North Asia, Grauer describes a sharp discontinuity in musical traditions of North America and Siberia:
“If the Americas had been populated directly and unproblematically via a Bering Strait land bridge in the manner usually presented, we would expect there to be a clear stylistic continuity between the music of the Paleosiberians of Siberia and the Amerindians of North America. But in fact there is no such continuity… In other words, between regions once connected by a land bridge, where we would expect to find continuity, we find discontinuity; and between regions separated by the vast reaches of the Pacific Ocean, where we would expect to find discontinuity, we find continuity.”
Grauer believes that musical evidence supports an early entry into the New World as part of the very first “coastal migration” out of Africa. While there’s little genetic or archaeological evidence for the popular “beachcomber migration” out of Africa, mtDNA data may provide a genetic counterpart to the unmistakable cultural similarities between the New World, Oceania and Sub-Saharan Africa. If mtDNA hg B and hg L0a2 are indeed related, this would push the age of the founding mutation 8281-8289d against the Neanderthal-attested 16189C background to earlier than the conventional (see Derenko et al’s phylogeny above) 50,000 YA postulated for hg B. It will also make African L lineages less of an outlier in the global mtDNA phylogeny and eliminate the bizarre situation whereby Pygmies have been underived for the past 100,000 years, while American Indians emerged only 12,000 years ago.



mtDNA L0a2 and B are not closely related. In fact, there is a large distance between L0 and all non-African sequences outside Africa. You can compare some B and L0 complete sequences here:
http://www.mitotool.org./database.html
Of course, there’s a large distance between certain lineages. All lineages are divergent from others to a certain degree. The question is whether 9-bp deletion is homoplastic in different parts of the globe, or it’s everywhere a signature of common descent. If it’s a single-event mutation, then the large divergence between L0a2 and B4’B5 in other loci suggests that African lineages may have mutated more rapidly than lineages outside of Africa. The advantage of seeing 9-bp deletion as a single-event mutation is that L0 is no more a deep outlier among human mtDNA lineages and consequently Khoisans, who have this haplogroup at high frequencies, did not diverge from other humans some 150,000 years ago. The latter date is 100,000 years earlier than the emergence of secure signs of modern human behavior globally. It’s rather preposterous to think that Khoisan have been in isolation from the rest of humanity for that long but then ended up having the same modern human language ability and cultural behavior as, say, the B4’B5 carriers in the Circumpacific region. In a word, all those sites at which L0a2 and B4’B5 are different are likely only “skin-deep,” while their sharing of 8281-8289d and 16223 (C) is the only thing that matters. If one finds that 8281-8289d and 16223 (C) are considered to be the only two mutations necessary to define hg B4’B5 qs a monophyletic clade, and these mutations are also present at L0a2 and T2f, then it seems legitimate to entertain the possibility that the 8281-8289d and 16223 (C) combo is more upstream than it’s usually thought to be.
You wrote:
“If it’s a single-event mutation, then the large divergence between L0a2 and B4′B5 in other loci suggests that African lineages may have mutated more rapidly than lineages outside of Africa.”
That can not be true either as there is also a large distance between L0 and African L3. Other deletions are recurrent as well. A 6 bp deletion (106-111d) in HVR2 comes to mind, 523- 524-, ‘C’ deletions around 16189 etc.
This is fair. I should’ve bracketed L3 out. Regarding recurrent deletions, it’s on a case-by-case basis. 106-111d is, as far as I know, a South American Indian specific mutation that occurs against the hg A2 and D1 backgrounds. It’s not found elsewhere (http://www.ncbi.nlm.nih.gov/pubmed/8964473) (but correct me if I’m wrong). What’s the hg D1 with 6-bp deletion state at np 16360? If it’s C16360T, then both loci are identical between the A2 and D1 lineages with 6-bp deletion. Plus one A2 sequence (#50 in Achilli et al. 2008) seems to have 6-bp insertion, which is unlikely. So, the data is far from clean. 6 bp deletion on D1 hasn’t been well-documented (only Merriwether et al. Am J Hum Gen 56, 1995, 812-813) and it’s phylogenetic importance is negligible compared to the importance of the 9-bp deletion that’s found against all possible backgrounds and geographies. It’s even found on hg B6, which is just one mutation step away from B4’B5. The argument for the 106-111d homoplasy originally used the 9-bp deletion as a supporting case (see Santos in Am. J. Hum. Genet. 57:195-196, 1995). Now, you’re using the 106-111d homoplasy as support for the 9-bp deletion homoplasy. It’s a circular argument.
[…] kind of “noise” (see also here on hg B and 9bp deletion) casts doubts on the robustness of existing mtDNA phylogenies. Tags: […]