Khoisans Are Genetically Admixed and Not Basal to Other Humans, Hadza Are Recently Admixed
In my last post I discussed the ways in which phylogenetic trees constructed on the basis of genetic data confound population relationships derived from common descent and from admixture. Unless admixture is taken into account, the sampled populations may end up in wrong places on the tree. I concluded the spot with the hypothesis that Khoisans and Pygmies “are likely products of waves of admixture in South and Central Africa.” The new paper by Joseph Pickrell et al. (2012) “The Genetic Prehistory of Southern Africa” (via Dienekes) confirms my insight on the theoretical level as well as on the level of application to Khoisan data. Pickrell et al. (2012) describe the way they filtered out “known admixture” from Khoisan populations:
“In the original TreeMix algorithm, one first builds the best-fitting tree of populations. However, this approach is not ideal if there are many admixed populations (as in our application here, where all of the Khoisan populations are admixed). To get around this, we allow for known admixture events to be incorporated into this tree-building step. Imagine that there are several populations that we think a priori might be unadmixed (in our applications, these are the Chimpanzee, Yoruba, Dinka, Europeans, and East Asians). We first build the best tree of these unadmixed populations using the standard TreeMix algorithm. Now assume we have an independent estimate of the admixture level of each Khoisan population, and imagine we know the source population for the mixture. To add a Khoisan population to the tree, for each existing branch in the tree, we put in a branch leading to the new population. We then force the known admixture event into the graph with a fixed weight, update the branch lengths, and store the likelihood of the graph. After testing all possible branches, we keep the maximum likelihood graph. We then try all possible nearest-neighbor interchanges to the topology of the graph (as in the original TreeMix algorithm), keeping the change only if it increases the likelihood. We do this for all populations. Finally, after adding all the populations with fixed admixture weights, we optimize the admixture weights, and attempt changes to the graph structure where the source populations for the admixture events are changed.”
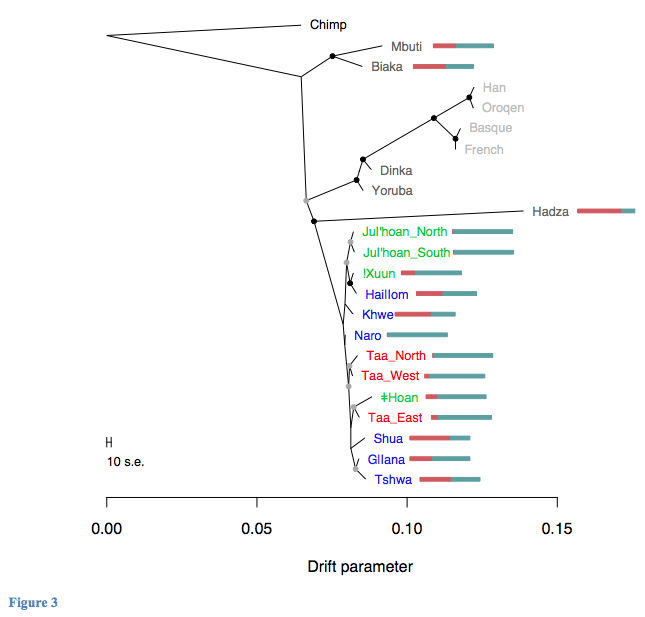
The phylogenetic tree of southern African populations adjusted for “known” admixture among Khoisan ended up looking like this (Fig. 3, Bantu-like ancestry is in red, non-Bantu-like ancestry in blue, with both “ancestries” being an umbrella term for diverse actual sources of gene flow):
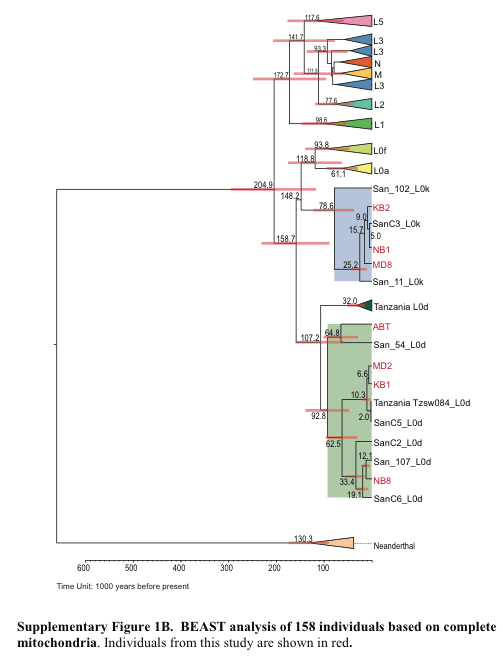
And now let’s compare this new tree with one of a myriad of trees that depicted Khoisans as a “basal human population.” As an example, I took an mtDNA tree from Schuster et al. (2010). “Complete Khoisan and Bantu Genomes from Southern Africa,” Nature 463: 943-947.
The surprising outcome of the comparison between an admixture-neutral and an admixture-corrected trees is that Khoisans are not a “basal population” anymore. They are further removed from the chimp-human node than Pygmies, Hadza as well as the branch that includes Dinka, Yoruba, Europeans and East Asians. This is true even for the least admixed Ju’hoan Bushmen.
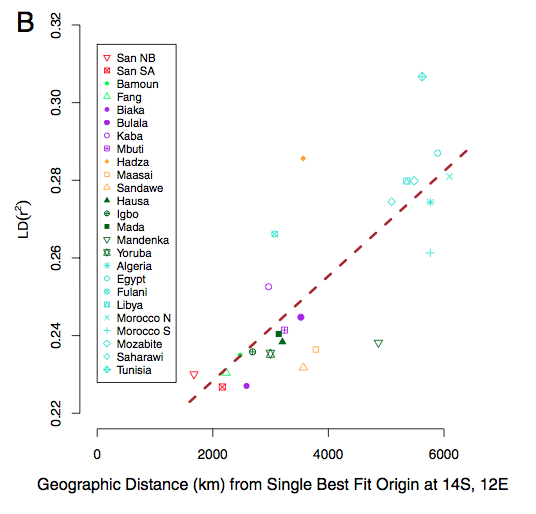
It’s noteworthy that Hadza show a large amount of Bantu-like ancestry. As I detailed in one of my web discussions, East African Hadza look like a genuine genetic isolate, possibly speaking the most divergent Khoisan language, having depressed heterzygosity values and representing the closest approximation to a Paleoafrican substratum among living African foraging populations. Pickrell et al.’s analysis confirms that they have a Khoisan component, that they form an outgroup to southern Khoisans and suggests that high linkage disequilibrium (LD) values reported from Hadza in Henn et al. (2011) (“Hunter-gatherer Genomic Diversity Suggests a Southern African Origin for Modern Humans,” PNAS 108 (13), Fig. 2B, see below) reflect not a recent bottleneck, but recent admixture with Bantu-like populations. (See more here on the cases when admixture is responsible for high LD.)
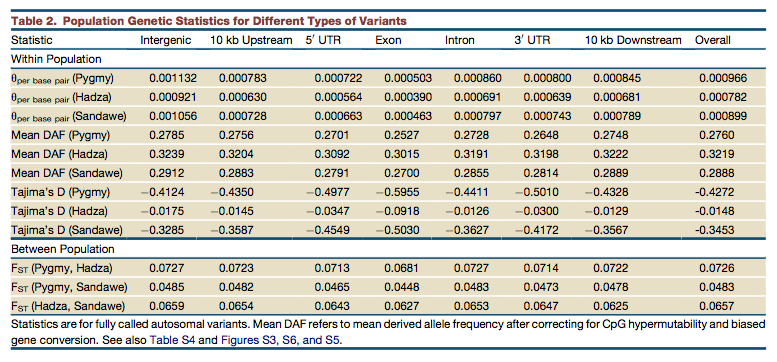
Lachance et al. (“Evolutionary History and Adaptation from High-Coverage Whole-Genome Sequences of Diverse African Hunter-Gatherers,” Cell 150, 457-469, 2012) confirmed the unique position of Hadza among African foragers. They write,
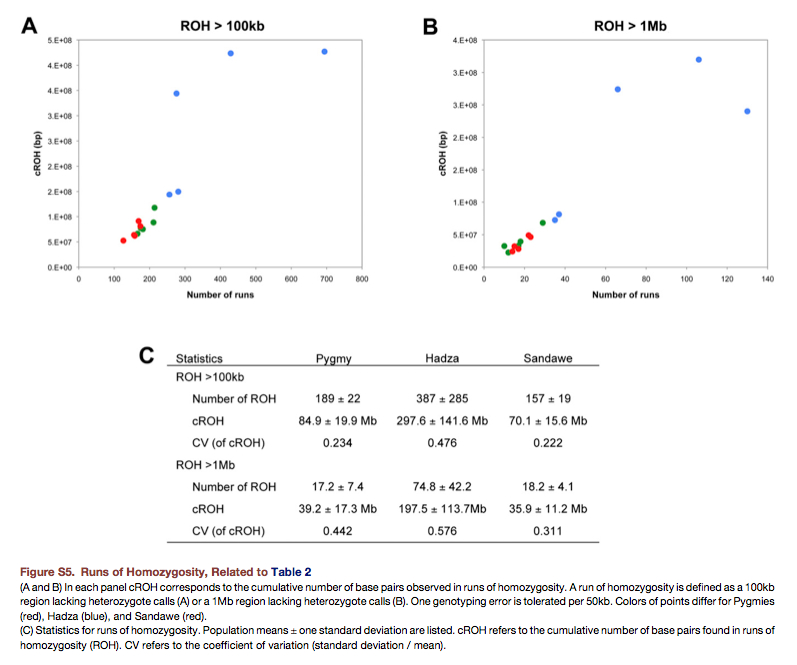
“Of the 15 hunter-gatherer genomes analyzed in this paper, the five genomes with the most runs of homozygosity all belong to the Hadza (Figure S5). Though some of these differences may be due to a population bottleneck in the Hadza (Henn et al., 2011), an additional cause may be cryptic inbreeding (Pember- ton et al., 2012), as indicated by the large variance in cumulative size of runs of homozygosity within the Hadza (Figure S5, see below on the right – G.D.). Indeed, cumulative runs of homozygosity in three Hadza genomes are more than double the size of other hunter-gatherers analyzed in this paper (Figure S5). Consistent with an historic bottleneck and/or inbreeding in the Hadza, we find that the proportion of polymorphic sites, as quantified by q, is lowest for the Hadza and highest for Pygmies (Table 2, see below on the left – G.D.). Depending on mutation rates, this trans- lates to effective population sizes of 11,300–25,700 (Pygmy), 9,200–20,900 (Hadza), and 10,600–24,000 individuals (Sandawe).”
This is exactly the kind of picture researchers usually obtain from American Indian populations. In both cases, long-term low effective population size, genetic drift and inbreeding (often associated with cross-cousin marriage marriage) are the explanations for the pattern.
The uniqueness of Hadza can also be seen in their cultural pattern. Victor Grauer gives the following assessment to Hadza music:
“To my knowledge no one has ever done a systematic study of Hadza music, so my impressions are based exclusively on this particular set of recordings, which might not be fully representative. On this basis, it would seem that Hadza singing, like Hadza autosomal markers, is highly distinctive — very beautiful indeed, but quite different from any other type of sub-Saharan African music with which I am acquainted. For one thing, the vocalizing on these CDs is almost exclusively in “social unison,” where all voices sing more or less in the same rhythm (albeit polyphonically, in a manner not unlike the parallel organum of the Medieval church) – while not unknown in sub-Saharan Africa, social unison is far less common there than call and response antiphony. Moreover, the Hadza tend to sing in relatively free, loosely coordinated, rhythms, and the rhythmic relation between the accompanying handclaps and the voices doesn’t appear to be very clearly coordinated — all of which is very unusual for Africa south of the Sahara, where rhythms and rhythmic coordination tend to be clearly defined and precise. While the existence of musical isolates is not unheard of in Africa, the unusual style of Hadza vocalizing is surprising and also puzzling, especially since they have so much else in common with Pygmies and Bushmen, whose musical style we could expect them to share.”
What remains to be seen is how admixture affects all the other African populations, including Pygmies and Bantu. They still appear as more “basal” than non-Africans, but it’s becoming increasingly clear that robust genetic phylogenies are only possible if they are grounded in a thorough admixture analysis. It appears that population geneticists are only now beginning to realize what has been a cornerstone of historical linguistics for more than a century. Effects of horizontal transmission (gene flow in the case of genetics and borrowing in the case of linguistics) as well as effects of homoplasy or convergence need to be sorted out prior to arriving at a realistic phylogeny. Pickrell et al.’s analysis should also serve as a cautionary tale to non-geneticists such as Victor Grauer and Alan Barnard (see more here) who try to use genetic trees as a model for modeling patterns of variation found in such cultural forms as music and kinship systems.