A Closer Look at Human and Neandertal Mitogenomes
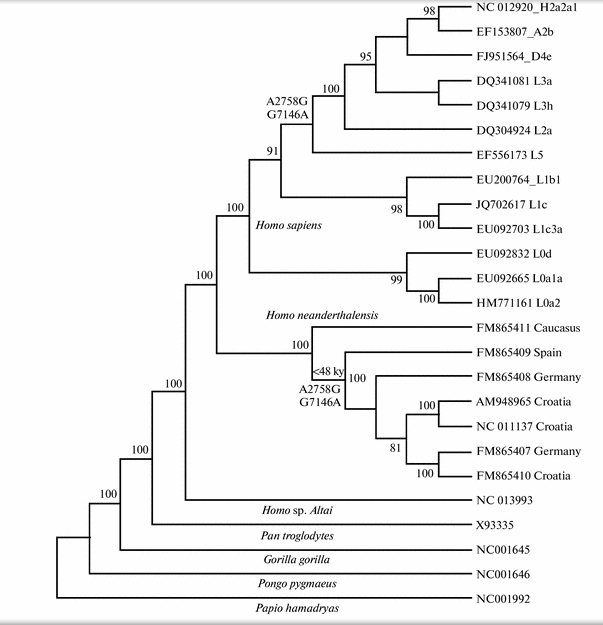
In one of my comments on Anthrogenica.com, I mistakenly denied that Behar et al. (2012) reported the two mutations A2758G and G7146A shared between late European Neandertals and the modern human L2’3’4’5’6 clade. As a reminder, A2758G and G7146A were highlighted in a recent paper by Malyarchuk as a possible evidence of hybridization with recombination between European Neandertals and modern humans belonging to the L2’3’4’5’6 clade (see phylogeny below).
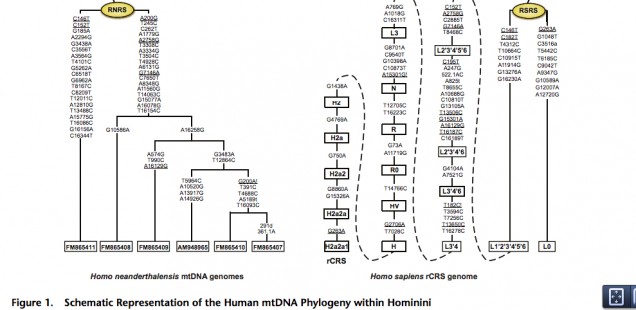
Behar et al. 2012 did this and in fact more. Let’s look closer at their tree (Fig. 1) and the associated table (Table S1 from Supplemental Material) showing the pattern of nucleotide sharing between the Homo sapiens reference sequence (RSRS) and the Neandertal reference sequence (RNRS).
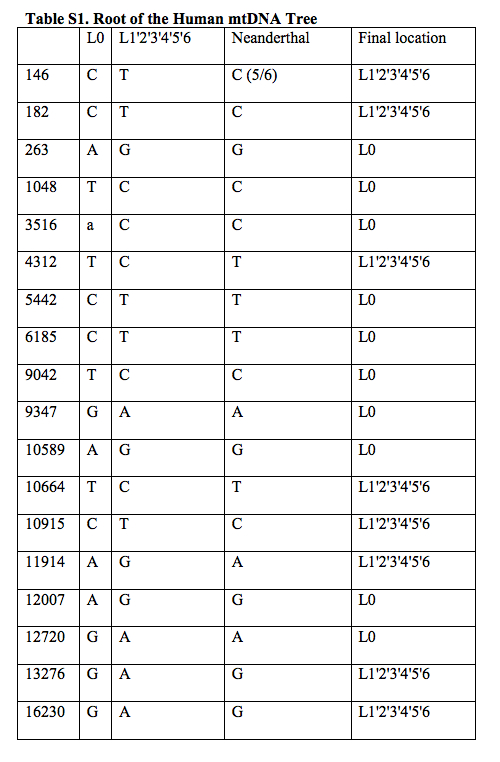 To begin with, L1’2’3’4’5’6 clade has fewer differences from RNRS than the L0 clade. L1’2’3’4’5’6 has 8 of those (C146T, C182T, T4312C, T10664C, C10915T, A11914G, G13276A, G16230A), while L0 has 10 (G263A, C1048T, C3516a, T5442C, T6185C, C9042T, A9347G, G10589A, G12007A, A12720G ). In addition, L1’2’3’4’5’6 shares mutation C146T with Mezmaiskaya (FM865411) bringing the count down to 7. The two mutations mentioned by Malyarchuk are located between the two nodes – L1’2’3’4’5’6 and L2’3’4’5’6 – on Behar’s tree. L2’3’4’5’6, therefore, has two more mutations in common with, now, the European branch of Neandertals (FM865408, FM865409, AM948965, FM865410, FM865407) bringing the count down to 5. Plus – this fact not mentioned by Malyarchuk – another mutation leading to L2’3’4’5’6, C152T, is found on Mezmaiskaya (FM865411), which forms the other Neandertal branch off of RNRS. We now have only 4 mutations separating the Neandertal sequences from Europe and the Caucasus from the modern human L2’3’4’5’6 clade. If we go further down the modern human mtDNA tree and “lose” African-specific L5 lineage, we see another match, namely A16129G, now between human L2’3’4 and Neandertal FM865409. As we “lose” another African-specific lineage, namely L2, and approach L3’4, we discover back-mutation T182C! that restores the original nucleotide to its ancestral state attested on RNRS. There are dozens of sites, of course, at which Neandertal and L3’4 lineages diverge, but it seems that L1’2’3’4’5’6 down to L3’4 has progressively more matches with a pool of Neandertal sequences than L0, although I may have missed them in L0 sub-branches.
To begin with, L1’2’3’4’5’6 clade has fewer differences from RNRS than the L0 clade. L1’2’3’4’5’6 has 8 of those (C146T, C182T, T4312C, T10664C, C10915T, A11914G, G13276A, G16230A), while L0 has 10 (G263A, C1048T, C3516a, T5442C, T6185C, C9042T, A9347G, G10589A, G12007A, A12720G ). In addition, L1’2’3’4’5’6 shares mutation C146T with Mezmaiskaya (FM865411) bringing the count down to 7. The two mutations mentioned by Malyarchuk are located between the two nodes – L1’2’3’4’5’6 and L2’3’4’5’6 – on Behar’s tree. L2’3’4’5’6, therefore, has two more mutations in common with, now, the European branch of Neandertals (FM865408, FM865409, AM948965, FM865410, FM865407) bringing the count down to 5. Plus – this fact not mentioned by Malyarchuk – another mutation leading to L2’3’4’5’6, C152T, is found on Mezmaiskaya (FM865411), which forms the other Neandertal branch off of RNRS. We now have only 4 mutations separating the Neandertal sequences from Europe and the Caucasus from the modern human L2’3’4’5’6 clade. If we go further down the modern human mtDNA tree and “lose” African-specific L5 lineage, we see another match, namely A16129G, now between human L2’3’4 and Neandertal FM865409. As we “lose” another African-specific lineage, namely L2, and approach L3’4, we discover back-mutation T182C! that restores the original nucleotide to its ancestral state attested on RNRS. There are dozens of sites, of course, at which Neandertal and L3’4 lineages diverge, but it seems that L1’2’3’4’5’6 down to L3’4 has progressively more matches with a pool of Neandertal sequences than L0, although I may have missed them in L0 sub-branches.