An Out-of-America Signal as Seen Through Human Regulatory Genes
PLoS Genet 9(4): e1003404. doi:10.1371/journal.pgen.1003404
Balancing Selection on a Regulatory Region Exhibiting Ancient Variation That Predates Human–Neandertal Divergence
Omer Gokcumen, Qihui Zhu, Lubbertus C. F. Mulder, Rebecca C. Iskow, Christian Austermann, Christopher D. Scharer, Towfique Raj, Jeremy M. Boss, Shamil Sunyaev, Alkes Price, Barbara Stranger,Viviana Simon, and Charles Lee.
Ancient population structure shaping contemporary genetic variation has been recently appreciated and has important implications regarding our understanding of the structure of modern human genomes. We identified a ~36-kb DNA segment in the human genome that displays an ancient substructure. The variation at this locus exists primarily as two highly divergent haplogroups. One of these haplogroups (the NE1 haplogroup) aligns with the Neandertal haplotype and contains a 4.6-kb deletion polymorphism in perfect linkage disequilibrium with 12 single nucleotide polymorphisms (SNPs) across diverse populations. The other haplogroup, which does not contain the 4.6-kb deletion, aligns with the chimpanzee haplotype and is likely ancestral. Africans have higher overall pairwise differences with the Neandertal haplotype than Eurasians do for this NE1 locus (p less than 10-15). Moreover, the nucleotide diversity at this locus is higher in Eurasians than in Africans. These results mimic signatures of recent Neandertal admixture contributing to this locus. However, an in-depth assessment of the variation in this region across multiple populations reveals that African NE1 haplotypes, albeit rare, harbor more sequence variation than NE1 haplotypes found in Europeans, indicating an ancient African origin of this haplogroup and refuting recent Neandertal admixture. Population genetic analyses of the SNPs within each of these haplogroups, along with genome-wide comparisons revealed significant FST (p = 0.00003) and positive Tajima’s D (p = 0.00285) statistics, pointing to non-neutral evolution of this locus. The NE1 locus harbors no protein-coding genes, but contains transcribed sequences as well as sequences with putative regulatory function based on bioinformatic predictions and in vitro experiments. We postulate that the variation observed at this locus predates Human–Neandertal divergence and is evolving under balancing selection, especially among European populations.
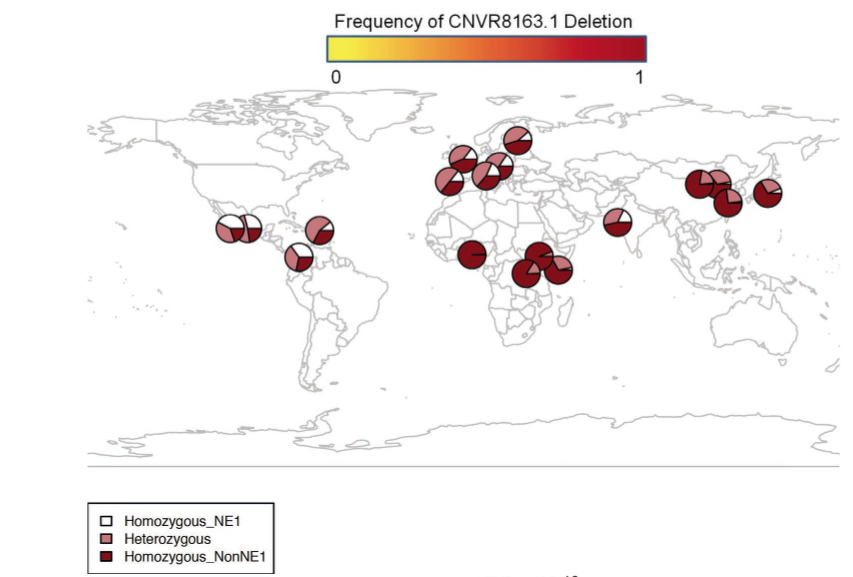
As the abstract indicates, Gokcumen et al. (2013) think they have advanced evidence for ancient African substructure that predates Human-Neandertal-Denisovan divergence and persists in Eurasian populations. The actual data is indeed interesting and deserves a broad discussion but the presentation of the data by Gokcumen et al. (2013) is biased against one continental population, namely American Indians. At the beginning, they do map out the worldwide distribution of the haplotypes in question (see below) and I will return to this data later.
But then American Indians are not featured in any of the subsequent calculations and interpretations and the worldwide conclusions are made on the basis of just YRI (Yoruba), CEU (European), CHB (Chinese) and JPT (Japanese) samples. For instance,
“For the majority of genomic loci, π is higher among YRI than among CEU (European ancestry) and CHB/JPT (Chinese/Japanese ancestry) populations. However, there is a marked increase in π among Eurasians, but not in YRI, for the NE1 locus especially around the regions flanking CNVR8163.1.”
“To identify potential gene targets of the putative regulatory sites within the NE1 locus, we performed a genome-wide cis– and trans– expression quantitative trait loci (eQTL) analysis in the three populations (CEU, CHB/JPT, YRI)…”
“We included in our analyses each probe that mapped to an Ensembl gene, but not to more than one Ensembl gene (Ensembl 49 NCBI Build 36) for probes in autosomal chromosomes. We excluded probes mapping to the X or Y chromosome as splitting the sample set to male and female cohorts would significantly reduce the power of our analysis. The resulting set of 21,800 probes was subjected to association analyses, corresponding to 18,226 unique autosomal Ensembl genes. We tested these associations with all of the SNP genotypes regardless of the haplogroup in 109 CEU, 162 CHB/JPT and 108 YRI samples located within the 36 kb region.”
“This LD block is evident in Eurasian (CEU and CHB/JPT) populations but is absent in the Yoruban (YRI) population…”
We already know that American Indian populations are regularly subjected to a sampling bias. One of these cases involves the innate immunity gene (OAS1), which, incidentally, Gokcumen et al. (2013) list among the regions showing the same pattern (as measured by π, LD, Tajima’s D and FST) suggestive of “archaic” admixture or substructure as the NE1 locus.
There seems to be a good reason for Gokcumen et al. (2013) to leave American Indian populations outside of their analysis – they would undermine their interpretation. As the above map of the distribution of the two haplotypes, NE1 and non-NE-1, demonstrates, American Indians show the world highest frequencies of homozygous NE1 haplotypes and the world highest frequencies of heterozygous (NE1/non-NE1) haplotypes. The NE1 haplotypes are precisely the ones that were detected in Neandertals and Denisovans.
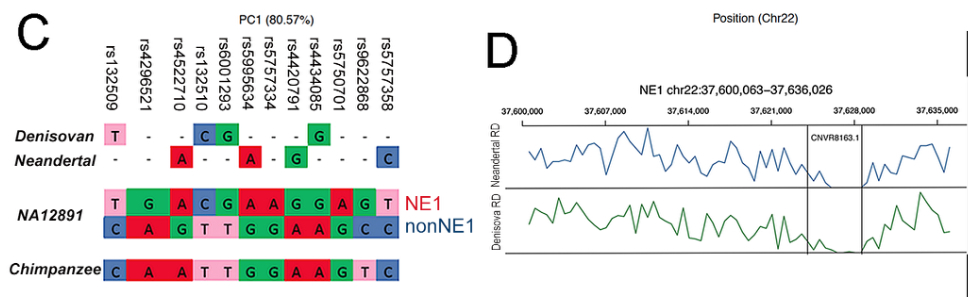
“Of the 12 SNPs that can be used to distinguish the NE1 and nonNE1 haplogroups, the SNPs that define the NE1 haplogroup aligned well with both the Neandertal and Denisovan orthologous sequences, whereas the chimpanzee consensus haplotype contain SNPs that are more similar to the nonNE1 haplogroup sequence (Figure 2C).”
“[R]ead-depth analyses of the Neandertal and Denisovan sequences across the CNVR8163.1 deletion interval supports the notion that this sequence is homozygously deleted in sequenced ancient hominins, but not in the chimpanzee reference sequence (Figure 2D).”
This is not an aberrant finding. The two Neandertals tested for blood group alleles showed blood group O (Lalueza-Fox et al. 2008. “Genetic Characterization of the ABO Blood Group in Neandertals,” BMC Evolutionary Biology 8: 342), which is most frequent among American Indians. The least derived B006 haplotype in the X chromosome’s dystrophin gene (ds44) was observed in modern humans and in Neandertals. In modern humans the frequency of B006 was the highest among North American Indians followed by Europeans. Other examples of Neandertal-Denisovan-Amerindian genetic similarities are discussed here.
Interestingly, just like in the case of B006, NE1 is most frequent in Amerindians followed by Europeans and South Asians. From the point of view of the conventional theory of the origin of Amerindians from East Asia, this fact is unexpected. But it’s fully consistent with the autosomal and mtDNA studies that identified “Amerindian admixture” in European populations. The NE1 case study suggests that “Amerindian admixture” and “Neandertal admixture” are different labels for the same phenomenon.
Now, neither Neandertals nor Denisovans have ever been found in the New World. This undermines the theory that modern humans admixed with these hominin species because one would expect to find the highest frequencies of an introgressed haplotype in the geographical area in which admixture took place. But the world highest frequencies of Neandertal-Denisovan haplotypes among Amerindians are equally inconsistent with the African substructure hypothesis presented by Gokcumen et al. (2013). While they conclude that
“the most parsimonious explanation for the observed variation at the NE1 locus is that the NE1/nonNE1 haplogroups arose after the human-chimpanzee common ancestor, but before the Human-Neandertal split in Africa. As such, the variation at the NE1 locus has persisted within ancient African substructure and later spread to non-African populations”
they are missing a key piece of the evidence – fossil DNA from Africa. NE1 haplotypes are ascertained in Eurasian hominins whose geographic range was likely adjacent to the New World, so if ancient substructure is at play here, it’s American and not African. The chimp-ascertained non-NE1 haplotype (the one without the deletion) is found in the New World, which suggests that this truly archaic genetic signature that predates the Neandertal-Denisovan-modern human split has survived there as well. It’s the more modern NE1 haplotype that shows a cline from the New World to Africa suggestive of a migration out-of-America leaving a clear trace on top of the undifferentiated chimp heritage.
Gokcumen et al. (2013) have identified a few interesting facts. First, contrary to the earlier accounts of “Neandertal admixture” in the human genome that failed to detect traces of it among African foragers, they found NE1 in 13% of their Mbuti Pygmy sample. This is precisely what out-of-America II predicts: modern humans with roots in a Eurasian hominin such as Neandertals or Denisovans colonized every remote corner of the world, including the African tropics. Second, they
“found that variation within African NE1 haplotypes is significantly higher than variation within Asian and European NE1 haplotypes (p<10−15)….”
But despite the higher variation within African NE1 haplotypes, the frequency of those haplotypes are the highest outside of Africa and, especially in America. This means that diversity is no indication of a population’s age. Plain simple. It’s likely that it shows the relaxation of a selective constraint in this region and the corresponding increase in mutation rate, which is something to be expected from a regulatory gene. Linkage disequilibrium (LD) metrics seem to support this interpretation (Amerindians again not represented):
“To understand the genomic composition upstream of the APOBEC3 locus, we first examined the phase I SNP data from the 1000 Genomes Project and identified an unusually strong linkage disequilibrium (LD) block spanning approximately 36 kb (NE1 locus, hg18 – chr22:37,600,063–37,636,026). This LD block is evident in Eurasian (CEU and CHB/JPT) populations but is absent in the Yoruban (YRI) population.”
The increase of genetic diversity as a result of the relaxation of selective constraints is something that is well described for some of modern humans’ companion species such as domesticated dogs and domesticated yaks following the domestication. A similar process must have affected their owners, too.