Encephalization, Fatty-Acid Metabolism and Modern Human Origins
American Journal of Human Genetics 90 (2012), 809–20. doi: 10.1016/j.ajhg.2012.03.014
Genetic Adaptation of Fatty-Acid Metabolism: A Human-Specific Haplotype Increasing the Biosynthesis of Long-Chain Omega-3 and Omega-6 Fatty Acids
Ameur, Adam, Stefan Enroth, Asa Johansson, Ghazal Zaboli, Wilmar Igl, Anna C.V. Johansson, Manuel A. Rivas, Mark J. Daly, Gerd Schmitz, Andrew A. Hicks, Thomas Meitinger, Lars Feuk, Cornelia van Duijn, Ben Oostra, Peter P. Pramstaller, Igor Rudan, Alan F. Wright, James F. Wilson, Harry Campbell, and Ulf Gyllensten.
Omega-3 and omega-6 long-chain polyunsaturated fatty acids (LC-PUFAs) are essential for the development and function of the human brain. They can be obtained directly from food, e.g., fish, or synthesized from precursor molecules found in vegetable oils. To determine the importance of genetic variability to fatty-acid biosynthesis, we studied FADS1 and FADS2, which encode rate-limiting enzymes for fatty-acid conversion. We performed genome-wide genotyping (n ¼ 5,652 individuals) and targeted resequencing (n ¼ 960 individuals) of the FADS region in five European population cohorts. We also analyzed available genomic data from human populations, archaic hominins, and more distant primates. Our results show that present-day humans have two common FADS haplotypes—defined by 28 closely linked SNPs across 38.9 kb—that differ dramatically in their ability to generate LC-PUFAs. No independent effects on FADS activity were seen for rare SNPs detected by targeted resequencing. The more efficient, evolutionarily derived haplotype appeared after the lineage split leading to modern humans and Neanderthals and shows evidence of positive selection. This human-specific haplotype increases the efficiency of synthesizing essential long-chain fatty acids from precursors and thereby might have provided an advantage in environments with limited access to dietary LC-PUFAs. In the modern world, this haplotype has been associated with lifestyle-related diseases, such as coronary artery disease.
Introduction
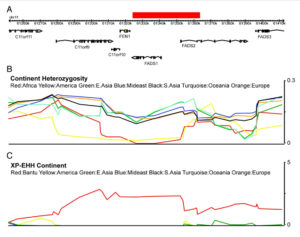
A map that encapsulates the critical findings of Ameur et al. (2012) provides a perfect illustration for out-of-America thinking.
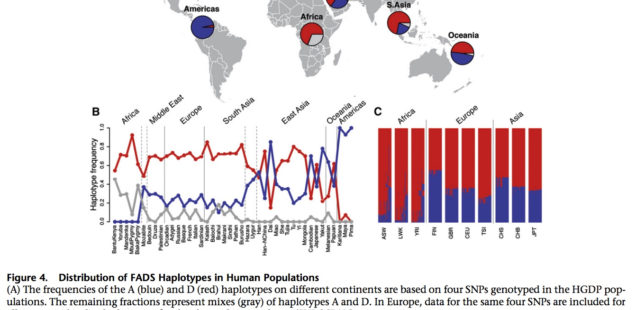
An ancestral allele is almost fixed in the New World, while a derived allele is almost fixed in Africa. Homozygosity at those loci is extreme in both America and Africa (see below, Ameur et al. 2012, Fig. S3).
Ameur et al. (2012) admit that this finding starkly “contrasts the more common pattern.” Harvard geneticist Iain Mathieson, too, acknowledged “strange patterns at FADS1 and FADS2.”
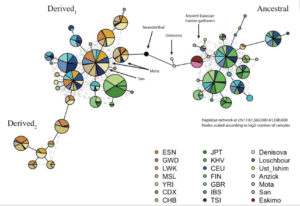
Mathieson constructed a haplotype tree in which derived haplotypes are concentrated in Europe and Africa, while ancestral haplotypes in East Asia, Papua New Guinea and America. Notably, some of the Sub-Saharan African haplotypes are most derived of all. (We have seen this picture before – the earliest enzyme morph analysis of human mtDNA by Johnson et al. 1983 generated a somewhat similar tree, see here.)
At the same, time FADS1 and FADS2 genes preserve the typical contrast between the two poles of modern human genetic variation – the Amerindian and the African. It’s the evolutionary placement of the two poles that is curiously reversed.
The importance of Ameur et al. (2012) for the study of modern human evolution is hard to overestimate. They analyzed two proximate genetic loci responsible for fatty-acid metabolism, which in turn is critical to human brain development and encephalizaton.
Encephalization and Human Evolution
Encephalization, or the increased proportion of the brain mass to the overall body mass, has been a hallmark of human biological evolution. In a stereotypical example, if one takes an African rhinoceros, its brain’s mass is just 0.1% of its body mass. In The Descent of Man, Darwin famously wrote that
“the large proportion which the size of man’s brain bears to his body, compared to the same proportion in the gorilla or orang, is closely connected with his mental powers.”
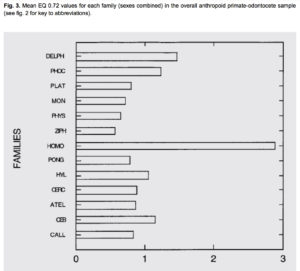
At that time, Darwin did not know that South American Cebidae (among which capuchins have recently been shown to manufacture hominin-grade stone tools) have an encephalization quotient that’s higher than that of Pongidae (see below, from Marino, L. “A Comparison of Encephalization between Odontocete Cetaceans and Anthropoid Primates.” In Brain, Behavior and Evolution, 1998).
While primates in general tend to violate the wider rule (well-attested in birds, for example) whereby pairbonding species tend to have bigger brains (see Schultz, S., and R. I. M. Dunbar. “The evolution of the social brain: anthropoid primates contrast with other vertebrates.” Proceedings of Biological Sciences 274 (2007): 2429–2436, Cebidae reinforce it: it’s a pair-bonding genus with a higher encephalization quotient than chimps, gorillas or orangutans. It’s another example of remarkable evolutionary convergence between Platyrrhines and modern humans that I have documented here. (But it’s precisely convergence, and not common descent, as Delphinidae, on the same chart, are even closer to humans on encephalization.)
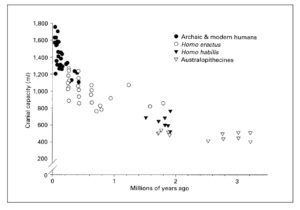
There’s ample paleontological evidence that encephalization nearly doubled between Homo erectus (EQ 2.38-2.44) and Homo sapiens (EQ 4.26). A corresponding revolution in cranial capacity is shown below (from Cordain, L. et al. “Fatty Acid Composition and Energy Density of Foods Available to African Hominids Evolutionary Implications for Human Brain Development.” In Nutrition and Fitness: Metabolic Studies in Health and Disease, edited by A. P. Simopoulos and K. N. Pavlou. Basel: Karger, 2001. P. 145).
Encephalization and Fatty-Acid Metabolism
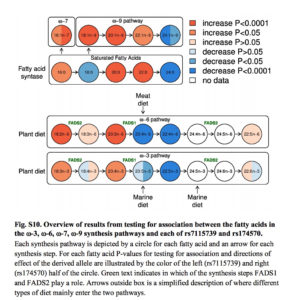
It’s also commonly believed that increasing encephalization in Homo was strongly dependent on the increasing intake of fatty acids. The human brain is 60% lipid and its functions heavily depend on long-chain polyunsaturated fatty acids (LC-PUFAs) that are relatively rare in nature. A progressively larger brain required progressively greater access to rich sources of the two LC-PUFAs – the omega-3 docosahexaenoic acid (DHA) and the omega-6 arachidonic acid (AA). They can either be absorbed directly from fish (rich in DHA) and land animals (rich in AA) or generated by the body from their precursors – middle-chain polyunsaturated fatty acids (MC-PUFAs), namely alpha-linolenic acid (ALA) and linoleic acid (LA) found in many vegetable oils. These two key enzymes (DHA and AA) are encoded by two genes, FADS1 and FADS2, respectively, located close to each other on chromosome 11. (Their proximity, by the way, is a result of a deletion of a DNA stretch that occurred in the human lineage.)
The process of enzyme generation and the dietary impact on it is shown below (from: Fumagalli, M. et al. “Greenlandic Inuit show genetic signatures of diet and climate adaptation.” Science 349 (2015), Suppl. Mat., p. 22).
What Ameur et al. (2012) found is that modern humans show two haplogroups associated with FADS1 and FADS2 – an ancestral one (A) shared with chimps, and a derived one (D) that’s uniquely ours. The derived one is conducive to the increased production of DHA and AA from their precursors, ALA and LA. In a sense, the ancestral allele is associated with a more natural situation when fatty acids are directly absorbed from fatty-acid-rich foods. The derived haplogroup emerged as a compensatory response to the paucity of fatty-acid-rich foods. Ameur et al. (2012: 809-10) write that
“mutations that increase the efficiency of converting the precursors ALA and LA to longer fatty acids are likely to be favored in environments with limited dietary access to these LC-PUFAs.”
Although its pathway for fatty-acid generation is more complex, haplogroup D is ultimately more efficient in producing DHA and AA. This can be seen in direct comparisons between African Americans and Caucasian Americans: the former have higher levels of LC-PUFAs than the latter (Mathias et al. 2012).
Amerindians are nearly homozygous on haplogroup A and Sub-Saharan Africans are nearly homozygous on haplotgroup D, with all other populations in-between. In Africa, foraging populations such as San and Pygmies are also haplogroup D, which indicates that the derived allele predates the splits within Sub-Saharan Africans and is not just a by-product of a Holocene shift to agriculture. Ameur et al. (2012) date the divergence of ancestral haplogroup A at 606,000 YBP, the emergence of haplogroup D in modern humans at 255,000 YBP. Mathias et al. (2012) date the onset of selection for haplogroup D in Africa to 85,000 YBP. All other things being equal, if the revolution in encephalization from Homo erectus to Homo sapiens and the corresponding increased intakes of fatty acids from local fish and land-animals started in Africa, modern African populations would be expected to be haplogroup A homozygous. Ameur et al. (2012, 817) write,
“Consequently, humans’ ability to more efficiently synthesize LC-PUFAs from their precursors might have played an important role their ability to survive in periods during which AA- and DHA-rich diets were not available. Haplotype D is likely to have been advantageous to humans living in environments with a limited access to these fatty acids, and this could explain the signature of positive selection seen for this haplotype in African populations.”
Why would Africans develop an adaptation to limited dietary sources of LC-PUFAs if ample dietary sources of LC-PUFAs are a pre-requisite for encephalization? This has been an unresolved issue in modern human origins. As (Mathias, R. A. et al. “Adaptive Evolution of the FADS Gene Cluster within Africa.” PLoS ONE 7 (2012): e44926 (2012)) point out,
“In fact, LC-PUFAs have long been considered to be the most limiting nutrients for neural growth and complexity during fetal and early childhood development, but they are not widely available in foods. Additionally, metabolic studies to date indicate that humans have little capacity to synthesize LC-PUFAs, especially DHA, from plant-based MC-PUFAs. Consequently, an unresolved question is how enough DHA and AA were acquired to support large brains and increases in brain complexity throughout human evolution. The prevailing view is that early human African populations obtained sufficient concentrations of LC-PUFAs to support brain evolution from DHA-enriched marine sources by living at the margins of lakes, rivers, or seashores in central and eastern Africa. But what facilitated the movement away from stable sources of DHA and expansions into a wide range of environmental (including arid) conditions?”
By an out-of-Africa logic, human populations leaving Africa would then encounter environments potentially less conducive to easy fatty-acid intake and haplogroup D would therefore evolve as an adaptation to the new dietary condition, or an “insurance” against unstable supplies of fatty-acid diets in new environments. The molecular facts, however, are the opposite from what we would expect under an out-of-Africa scenario.
Per Ameur et al, (2012), the New World has provided Amerindians with a rich source of fatty-acid-rich diets:
“The low frequency of haplotype D in the Native American populations included in HGDP indicates that this haplotype might have been lost because of a bottleneck effect in the colonization of the American continent, possibly in combination with relaxation of the selective pressure as a result of a diet higher in essential LC-PUFAs.”
So, the New World does satisfy the environmental conditions for human encephalization, while Africa does not. The African situation is derived, while the New World situation is ancestral, both environmentally and genetically, which solves the lingering problem in the origin and maintenance of fatty-acid intake in the course of human encephalization.
Hominin and ancient Eurasian DNA (see below, from Matheson, 2015) add more questions to the unusual global distributions of LC-PUFAs-coding alleles.
Within the pool of ancient Amerindian and Eurasian samples, Anzick (12,500 YBP, Paleoindian foragers), Saqqaq (4,000 YBP, Paleo-Eskimo foragers) and Ust’-Ishim (45,000 YBP, East Eurasian foragers) are haplogroup A homozygous (if haplogroup A introgressed into modern humans from the archaics, we would have found Ust’-Ishim to be heterozygous for A and D), Loschbour (8,000 YBP, West Eurasian foragers) has a transition at rs174551, just like Stuttgart (West Eurasian farmers), but Stuttgart is also heterozygous across all the other sites. In Africa, the ancient Mota Cave specimen (4,500 YBP, foragers) has a unique molecular signature comprised of ancestral homozygous, heterozygous and derived homozygous alleles. It appears that mutations away from the ancestral haplogroup have been happening independently and at different times in Eurasian hominins, West Eurasians and Africans. In West Eurasia, the 80% dominance of the derived haplogroup came about very late. At the same time, 50% of East Asians, most Papuans and Australian aborigines and virtually all Amerindians did not inherit any of those genetic changes. Within modern human samples, the older the specimen and the closer it is to America geographically, the more likely it is to be haplogroup A, just like most modern Amerindians are. Selective pressures coming from agricultural and pastoralist plant-rich (meat-and fish-depressed) diets in Europe could play a role in its rapid spread (see below, from Mathieson et al. “Genome-wide patterns of selection in 230 ancient Eurasians.” Nature 528 (2015), 499-503), but agriculture must have had only a local, moderating effect on the evolution from haplogroup A to haplogroup D considering that agricultural populations in America seem to be haplogroup A, while African foragers are haplogroup D.
Surprisingly, Neandertal and Denisovan haplotypes are more derived than human haplogroup A. In Mathieson’s haplotype tree (see below), they are depicted as intermediary between ancestral and derived human haplogroups: Devisovan is closer to ancestral lineages and Neandertal is closer to derived lineages.
Most recently (Buckley, M. T. et al. “Selection on the FADS region in Europeans.” Biorxiv, 2016, Suppl. Inf., p. 8) built a tree for the critical FASD SNP, namely rs174546, which confirmed the global distribution of ancestral vs. derived haplogroups and clarified the position of Denisovan vs. Altai Neandertal haplotypes. Human haplogroup A is intermediary between Denisovan and Neandertal haplotypes, with Denisovan being an outlier in the whole tree (see below).
Strangely enough, Ameur et al.’s phylogeny does not reflect the fact that the clade formed by Saqqaq and Australian aboriginal sequences is closer to the chimp sequence than Denisovan or human haplogroup D sequences (see below).
But regardless of the tree topology, the data excludes the possibility that haplogroup A came to modern human gene pool through “archaic admixture.” So, another tenet of most recent out-of-Africa thinking is disproved by the data. But the evidence at hand is consistent with one of the predictions of out-of-America II, namely that modern humans evolved from a Eurasian hominin species related to Neandertals and Denisovans. This ancestral Eurasian species would be haplogroup A. Amerindians, as an early isolate, have maintained it better than any other human population. As human populations expanded out of America, the consistency of DHA- and AA-rich diets’ availability faltered and a genetic mechanism to replenish fatty acids through the biosynthesis of their precursors kicked in. An interpretation whereby haplogroup D emerged in response to new niche colonization is consistent with the conclusion reached by Mathias et al. (2012):
“it is likely that suddenly having the capacity to more efficiently convert plant-based MC-PUFA to LC-PUFAs would have been an important advantage that would have facilitated expansion and movement into a variety of ecological locations.”
This seems to be the most parsimonious interpretation of the evidence: from the New World into Eurasia and, finally, Africa.