American Indians, Neanderthals and Denisovans: Insights from PCA Views
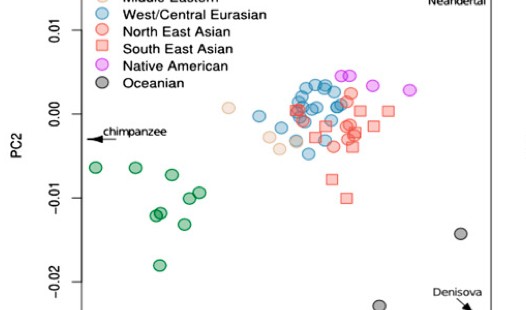
Dienekes posted a SNP PCA showing the relative position of a sample of modern human populations from the Harvard HGDP along the axes formed by Chimpanzees, Denisovans and Neanderthals. On the broad-view PCA, the red dot indicates Chimpanzees, the green dot Neanderthals and the blue dot Denisovans. In accordance with earlier studies, in the zoom-in version of the PCA, Eurasians shift toward Neanderthals, Papuans toward Denisovans, while Africans and, especially, San look closer to Chimpanzees.
Among Eurasians, Lahu and Naxi, two Sino-Tibetan populations from China, are the closest to Neanderthals. This is, by itself, is surprising, as Neanderthals must have had more extensive contact with Europeans, not East Asians. American Indians again are not represented in the sample. But if we look at another SNP PCA from Skoglund & Jakobsson’s “Archaic Human Ancestry in East Asia” (2011), we’ll see that it’s American Indians that are noticeably closer to Neanderthals.
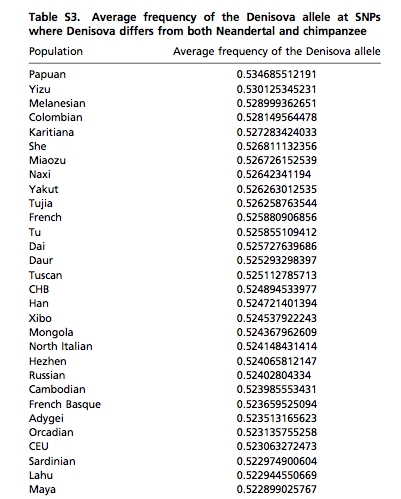
Where the Harvard HGDP sample failed, Skoglund & Jakobsson’s sample encompassing HapMap 3, Chicago HGDP and Finnish HapMap data identified the position of American Indians in PC1 and PC2. If American Indians were part of the sample from which Dienekes derived his PCA, they would have fallen in the space between Lahu/Naxi and Neanderthals. Skoglund & Jakobsson’s PCA also suggests some proximity between American Indians and Denisovans. This proximity comes out clearly in their Table S3 (see below in slightly shortened form), where Colombian and Karitiana are right next to Melanesians and ahead of Naxi in terms of frequency of the Denisova allele. Maya and Lahu are further down below but both are ahead of most West Asians, South Asians and West Eurasians. It appears that all easternmost populations, whether Melanesians, East Asians or South American Indians, are shifted closer to the Denisovan pole, while all of American Indians are shifted closer to Neanderthals.
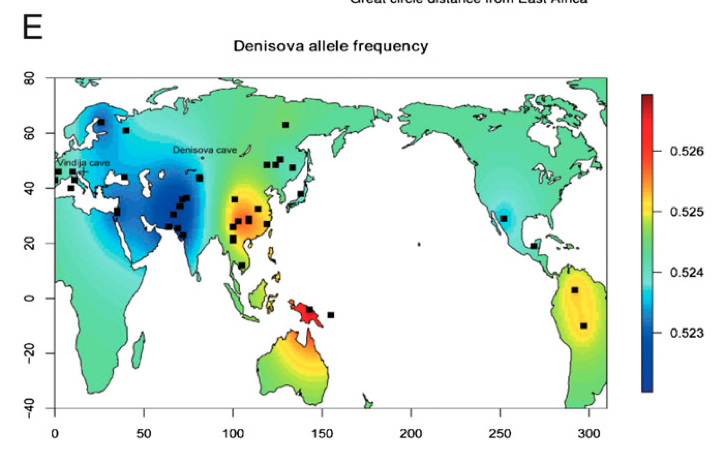
This is confirmed on Skoglund & Jakobsson’s map of the distribution of the Denisova allele in modern human populations.
The gradient connects Sahul, East Asia and America as the regions with most pronounced Denisovan admixture with West Eurasia and Africa (no Denisovan admixture). On Dienekes’s PCA some African populations seem to be shifting slightly in the direction of the Denisovan pole, as some of Dienekes’s readers noted in the Comments. It seems likely that just like Neanderthal admixture is apparently present in the Yoruba, Denisovan admixture may eventually be detected in some Sub-Saharan Africans at some of the lowest frequencies in the world.
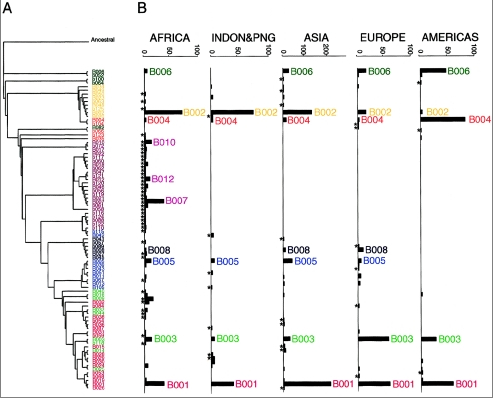
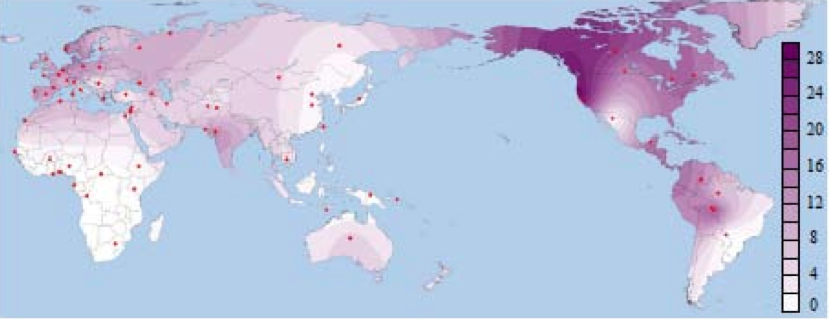
It’s interesting to compare the distribution of Denisovan “admixture” with the distribution of Neanderthal “admixture” in the polymorphisms observed in human X chromosome’s dystrophin gene (ds44). The least derived B006 haplotype was observed in modern humans and in Neanderthals. In modern humans the frequency of B006 was the highest among North American Indians followed by Europeans (see Ziętkiewicz et al. Haplotypes in the Dystrophin DNA Segment Point to a Mosaic Origin of Modern Human Diversity. Am J Hum Genet 2003 November; 73 (5): 994–1015). American Indians also showed other near-basal haplotypes B052, B064 suggesting that the conservation of B006 among them is part of a systemic phenomenon. Applying a formula of sequence divergence to estimate the age of B006 on the Americas, Austin Whittall obtained the figure of 188,000 years, which is roughly the same time (pending molecular clock optimizations) as the emergence of “anatomically modern humans” in the African archaeological record.
B006 has not been reported from the southern areas of Sub-Saharan Africa but one copy was observed in Ethiopia and four in Burkina Faso suggesting gene flow from the north. A map of the worldwide distribution of B006 is shown below (from Yotova et al. “An X-Linked Haplotype of Neandertal Origin Is Present Among All Non-African Populations,” Mol. Biol. Evol. 28 (7), 2011).
Similarities between the New World and Melanesia/Papua New Guinea also transpired in a large-scale analysis of worldwide microsatellite variation by Sarah Tishkoff’s lab.
“The ratio of variance and heterozygosity is the largest in Native American and Papuan and Melanesian populations followed by East Asians, all with values greater than one, intermediate in most Europeans, Middle Easterners, and Indians, with values near one, and with values less than one in most African populations and a few Middle Eastern and European populations” (Tishkoff et al. “The Genetic Structure and History of Africans and African Americans,” Science 324 (5930), 2009, Suppl. Mat., p. 10).
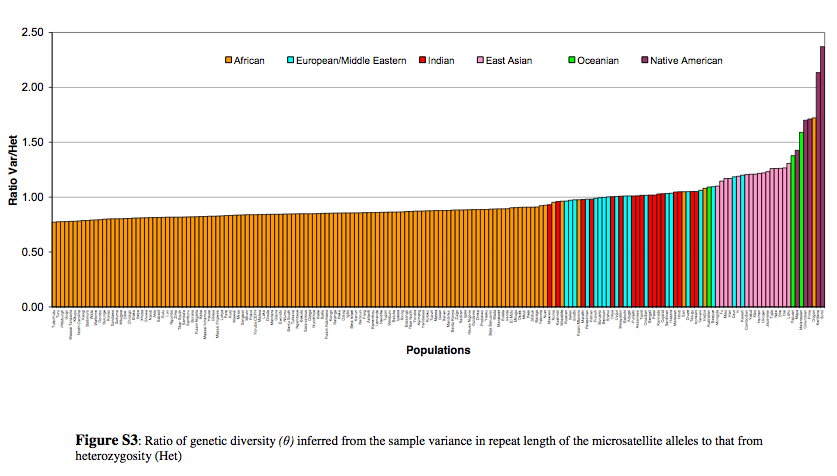 This means that American Indians are Melanesians/Papuans are small populations subject to genetic drift. Tishkoff et al. believe that the highest ratio of variance to heterozygosity also means that American Indians and Papuans/Melanesians went through “a strong bottleneck followed by a recent and rapid expansion,” but the recency of this event is cast into doubt given the “archaic admixture” evidence. Most interestingly, African hunter-gatherers, Hadza and San, have the lowest heterozygosity values among all African populations (Tishkoff et al., Suppl. Mat., p. 10), and on Tishkoff’s Fig. S3 (see above) they are noticeably shifted rightward in the direction of American Indians and Melanesians/Papuans. Tishkoff et al. attribute this unusual position of African foragers to their “stable small population sizes” (p. 10-11) and not to their recency. What Tishkoff et al.’s data seems to show is that American Indians and Melanesians/Papuans preserve an ancient population structure that’s also the ancient population structure of the Hadza and the San in Africa, but in Africa it’s overlaid by a thick stratum of agricultural and nomadic cattle-breeding diversity.
This means that American Indians are Melanesians/Papuans are small populations subject to genetic drift. Tishkoff et al. believe that the highest ratio of variance to heterozygosity also means that American Indians and Papuans/Melanesians went through “a strong bottleneck followed by a recent and rapid expansion,” but the recency of this event is cast into doubt given the “archaic admixture” evidence. Most interestingly, African hunter-gatherers, Hadza and San, have the lowest heterozygosity values among all African populations (Tishkoff et al., Suppl. Mat., p. 10), and on Tishkoff’s Fig. S3 (see above) they are noticeably shifted rightward in the direction of American Indians and Melanesians/Papuans. Tishkoff et al. attribute this unusual position of African foragers to their “stable small population sizes” (p. 10-11) and not to their recency. What Tishkoff et al.’s data seems to show is that American Indians and Melanesians/Papuans preserve an ancient population structure that’s also the ancient population structure of the Hadza and the San in Africa, but in Africa it’s overlaid by a thick stratum of agricultural and nomadic cattle-breeding diversity.
Going back to “archaic admixture,” higher frequencies of Neanderthal alleles among North American Indians and slightly lower frequencies of Denisovan allelles in South American Indians are very unexpected and don’t fit the “archaic admixture” theory, as there were no Neanderthals or Denisovans in America. There are ways to dismiss the inconvenient facts by attributing them to “ascertainment bias,” “false positives,” or “genetic drift.” But by the same token the proximity of the San to the Chimpanzee pole can be explained away without inferring that the San are basal to all other human populations. Science needs to have a strong theory to account for anomalous facts. That’s when other disciplines beside genetics come in handy. It may not be a coincidence that the signs of “archaic admixture” are the highest precisely where linguistic diversity is also the highest, namely in the New World and Papua New Guinea. It’s precisely the New World and the Sahul that Johanna Nichols (Linguistic Diversity in Space and Time, 1992) classified as linguistic “residual zones” that preserve grammatical structures suggestive of what the earliest human language may have been like. Ethnologists and ethnomusicologists find striking similarities between South America and Papua New Guinea in myths, rituals and music (see, e.g., the collection of essays Gender in Amazonia and Melanesia, edited by Thomas Gregor and Donald Tuzin, its review my folklorist Yuri Berezkin in Latin American Indian Literatures Journal 18 (2002), or Victor Grauer’s description of musical parallels). Haploid genetics (mtDNA and Y-DNA) hasn’t matched the unmistakable cultural and linguistic similarities between the New World and Papua New Guinea with specific haplotypes shared between the two regions. Neither has it matched the discovery of “archaic admixture” with similar “archaic” mtDNA and Y-DNA haplotypes. But haploid systems are subjected to stronger genetic drift and may have simply lost traces of an ancient allelic pattern connecting the New World and Papua New Guinea. If this is the case, then the patterns observed in cultural and linguistic data may be more conservative than some of the patterns detected in haploid genetics.
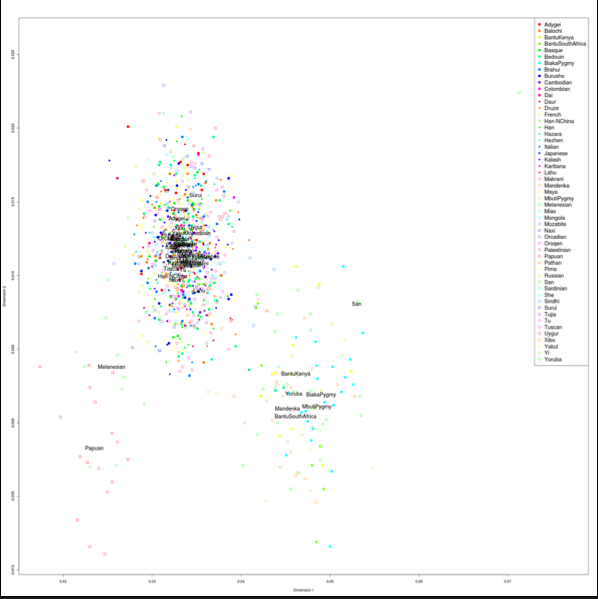
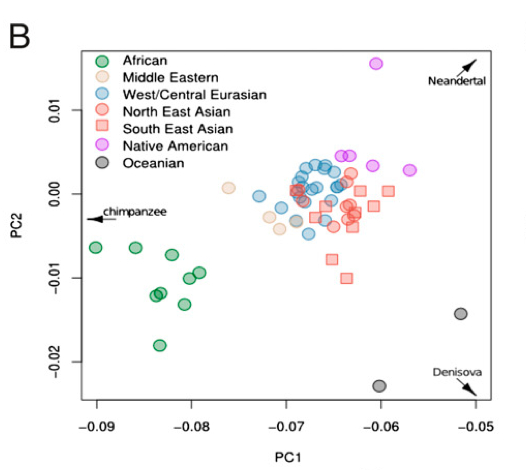


[…] associated PCA is very similar to the one that lacked American Indians. Neanderthals are bottom left, Denisovans are top […]
[…] Here’s a scientfic article that states that native American Indians have more Neanderthal DNA in them than other human populations: Neanderthal DNA in native Americans […]
The following comment supports An American Wellspring or Out of America I with no admixture from Denisova… also see the supplement last graph for negative values pointing to ancestral position for Amerindians. They are nodal for all genetic conditions including archaeologically, as pre-Clovis sites have nothing to do with Old World Paleolithic systems, being, rather, the signature of Ancient Modern Humans (using an outdated definition that needs to be revisited (vs archaic modern humans), original pre-Upper Paleolithic behavior/subsistence strategy.
“The apparent absence of Denisova ancestry in Native Americans in our study could be influenced by the biased affinity to the Neandertal genome that is expected because of ascertainment bias and genetic drift, but analyses of unascertained low-coverage shotgun sequence data from a single Native American individual resulted in a similar conclusion (24). If absent in America, the Denisova component must have appeared in the ancestors of East Asians after their divergence from Native Americans, which has been dated to ∼14–30 kya (58, 60, 61). from above Skoglund & Jakobsson’s “Archaic Human Ancestry in East Asia” (2011)”
Why are humans so spread out on the left-right axis? Chimps are so much farther than humans than the other species, so it’s like showing the left-right axis in miles and the north south in feet. Would random DNA do the same thing as chimp DNA?
Also, why not just plot the human-neanderthal distance vs the human-devension distance? No more arbitrary outgroups and no more “why are they shifted toward chimps?” confusion.
Read with interest this study, for as a lay-person who recently got the results of her National Geographic 2.0 Geome test results back, I was struck by the fact that I was listed as having 11% American Indian in me and 3.4% Denisovan. I could not for the life of me see where that percentage could have happened, and this study confirms the great impossibility of it having come from the American Indian side. I then remembered reading that several members of my extinct tribe (Maidu) had interbred with some native Hawaiians who had been brought over to work in the sugar fields here in California some time ago. That would definitely explain the influence of the Denisovan DNA in my book. All my other percentages dealt with my being Norther European.
[…] Os Homo sapiens – espécie exótica e invasora suprema – ao encontrar estes outros humanos fizeram o que continuariam fazendo depois: sexo e conquista. O resultado é que 1% a 4% do DNA das populações não africanas atuais é de origem Neandertal, enquanto até 6% do genoma dos nativos melanésios é Denisova.Alelos desta origem também aparecem no genoma de populações no sudeste asiático e de índios americanos. […]
My Genographic Project results show the same 3.4% Denisovan as Robertson’s with no Native American admixture. My take on this is that the Eurasian population should be thought of as a unit–not subdivided into Neandertal, Denisovan, and the like–but with a range of variation that includes the traits attributed to both. We need to remember that our Denisovan sample is limited to a single individual. I’ve seen no convincing evidence that would suggest that gene flow from Africa and across Eurasia was ever interrupted long enough for us to be talking about separate species.
Another point that must be borne in mind when looking at the graph that shows African populations as being closer to chimps is that both occupy African environments, which would tend to expose them to the same selective pressures, thus tending to preserve in human populations those alleles most fitted for those environments, while humans who moved out of Africa would be exposed to very different sets of selective pressures.
Thanks Ron. Welcome! We can’t use similar environmental pressure as a proxy for common descent. I’d say that the proximity of Africans to chimps speaks to the possibility of convergent evolution that kicked in after Africans had migrated to Africa from Eurasia.