The Effect of Long-Term Endogamy on Identity-By-Descent
PLoS ONE 7(4): e34267. doi:10.1371/journal.pone.0034267
Cryptic Distant Relatives Are Common in Both Isolated and Cosmopolitan Genetic Samples
Henn, Brenna M., Lawrence Hon, J. Michael Macpherson, Nick Eriksson, Serge Saxonov, Itsik Pe’er, Joanna L. Mountain.
Although a few hundred single nucleotide polymorphisms (SNPs) suffice to infer close familial relationships, high density genome-wide SNP data make possible the inference of more distant relationships such as 2nd to 9th cousinships. In order to characterize the relationship between genetic similarity and degree of kinship given a timeframe of 100–300 years, we analyzed the sharing of DNA inferred to be identical by descent (IBD) in a subset of individuals from the 23andMe customer database (n = 22,757) and from the Human Genome Diversity Panel (HGDP-CEPH, n = 952). With data from 121 populations, we show that the average amount of DNA shared IBD in most ethnolinguistically-defined populations, for example Native American groups, Finns and Ashkenazi Jews, differs from continentally-defined populations by several orders of magnitude. Via extensive pedigree-based simulations, we determined bounds for predicted degrees of relationship given the amount of genomic IBD sharing in both endogamous and ‘unrelated’ population samples. Using these bounds as a guide, we detected tens of thousands of 2nd to 9th degree cousin pairs within a heterogenous set of 5,000 Europeans. The ubiquity of distant relatives, detected via IBD segments, in both ethnolinguistic populations and in large ‘unrelated’ populations samples has important implications for genetic genealogy, forensics and genotype/phenotype mapping studies.
Link (Free Text PDF)
Flanked by two geneticists whom I know personally from my years at Stanford, Brenna Henn and Joanna Mountain (now both affiliated with 23andMe), this paper puts population genetics in close relationship with anthropology’s two all-time favorites – reflexivity and kinship studies. Similarly to another recent paper by Hunley et al. (2011) that suggested that higher levels of genetic diversity in North America over Central and, especially, South America may have been caused not by ancient bottlenecks accompanying the colonization of the Americas from north to south but by the varying degree of European gene flow into American Indian tribes in recent centuries, Henn et al. observe that high levels of identity-of-descent (IBD) in genetic samples taken from both small isolated and large cosmopolitan populations may come from cryptic consanguinity. Since the inception of the reflexive turn in anthropology in the late 1960s, the discipline has demonstrated heightened attention to the most recent historical processes as well as to the immediate context of the production of scientific knowledge. Positivist science tends to naively interpret every signal that comes its way as representative of either an ancient pattern or a general evolutionary regularity, as if these patterns and regularities are somehow served up by Mother Nature to reward scientists for their painstaking labor of finding answers to fundamental human questions. If modern African populations are genetically diverse and modern American Indian populations are genetically homogeneous, it’s because African populations are very old and American Indian populations are very young. If South American Indians are less diverse than North American Indians and North American Indians are less diverse than East Asians, it’s because their genomes maintain the signal of that epochal colonization of America from Siberia that Father Jose de Acosta was talking about.
Papers such as Hunley et al.’s and, now, Henn et al.’s deal a blow to these simplistic models. To quote Henn et al. at length,
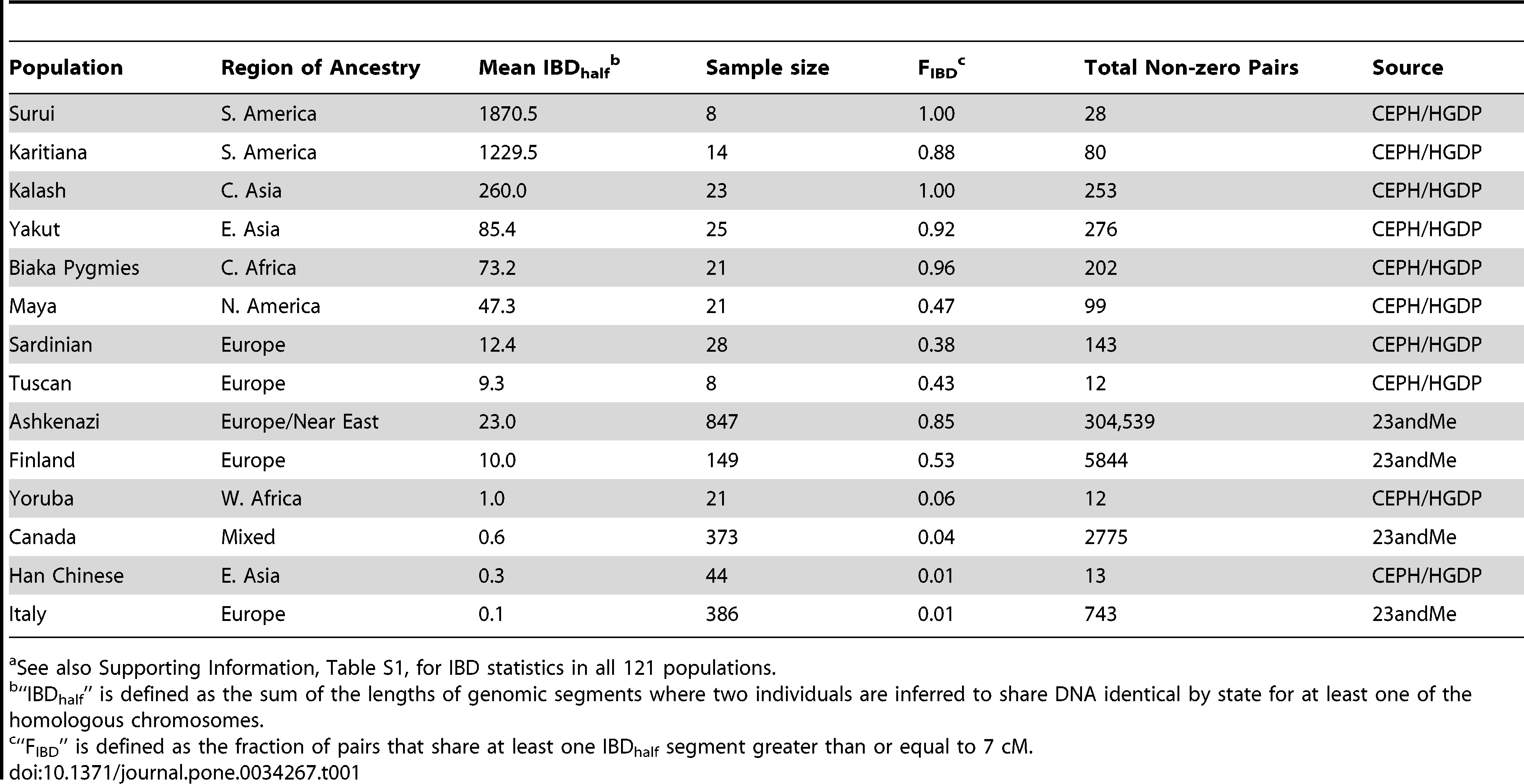
“The characterization of genomic similarity across samples of the HGDP-CEPH collection has revealed about 100 cryptically related or duplicate samples. Prior analyses focused on identifying familial relationships such as sibships, parent-offspring pairs, etc. (i.e., 1st–3rd degree relatives). We extended this analysis through pairwise IBD comparisons and characterized pairs of individuals who are related as 1st through 9th cousins within each HGDP population. When datasets include samples from populations where many of the individuals are closely related, analyses of population structure tend to cluster these populations more discretely than populations with greater genetic variation. For example, previous analyses of genome-wide microsatellite data identified the Kalash of Pakistan as a distinct global population in a STRUCTURE analysis of HGDP, k = 6. Our pairwise IBD metrics suggest an explanation for this result; the Kalash share on average 260 cM IBDhalf and thus any given pair of individuals is the genomic equivalent of second cousins. In both microsatellite and SNP genotype-based STRUCTURE plots, Native American HGDP populations emerged as a population subset at k = 4. Our IBDhalf estimates for the Native Americans are consistent with an interpretation that the majority of individuals within these populations are related as the genomic equivalent of 2nd cousins or closer. We note that although putative close relatives were removed with the HGDP952 dataset, our 700 cM cutoff employed for the 23andMe dataset would have removed most pairs in the Native Americans populations (though not from most HGDP groups). Populations such as the Karitiana or Pima display levels of mean IBDhalf that are 3-4 orders of magnitude higher than many HGDP Asian populations such as the Han, Japanese, Mongolians. As such, population genetic analyses of the HGDP-CEPH Native Americans will need to account for the IBD derived from their elevated levels of historic endogamy or uneven population sampling. The high levels of mean IBDhalf and FIBD of many of the 121 globally distributed populations we analyzed are consistent with substantial structure among populations within continents. On the other hand, populations that were sampled randomly across a wide ethnolinguistic and geographic space (i.e., 23andMe European and Asian continental samples) have very low estimates of mean IBDhalf. The elevated amount of IBDhalf in the majority of ethnolinguistic populations from HGDP-CEPH, and in some 23andMe sub-continental samples, compared to that of samples of general European or Asian ancestry is indicative of small effective population size, often reflecting endogamy.”
Earlier this year, Brenna Henn reported on a high IBD index in the Tunisian sample likely caused by the prevalence of first- and second-cousin marriage. The special position of South American Indian tribes, Surui and Karitiana, are highlighted in the following sample of worldwide populations. (Pima and Colombian natives that are not shown in the table below fall between Karitiana and Kalash.) Note the 20-fold difference in Mean IBDhalf between Surui and Biaka Pygmies. There’s an interesting parallelism between zones with high IBD (America, parts of Asia, Melanesia, African foragers) and low IBD (Eurasia) and Johanna Nichols‘s “residual” and “spread” zones defined on the basis of frequencies of grammatical markers.
This is exactly the pattern observed across virtually all genetic systems – American Indians are extreme outliers. However, Henn et al. don’t follow the usual interpretative path to explain the special position of American Indians as the function of a bottleneck after a recent separation from East Asians. Instead, they argue that South American Indians are the populations most affected by long-term endogamous practices, which naturally create samples with a large proportion of cryptic relatives. High prevalence of IBD in human populations is the direct molecular consequence of the social structure based on what anthropologists call “symmetric-prescriptive alliance,” “elementary structures of kinship,” “two-section, “two-line,” or “Dravidian-Kariera-Amazonian.” These systems, which anthropologists from Claude Levi-Strauss to Nick Allen postulated as ancestral to all other systems of kinship and marriage, are found at high frequency and in their purest forms in Amazonia. (See, most recently, Hornborg, Alf F. “Opposition, Hierarchy and Gender in Aboriginal South America: Linguistic and Architectural Homologies,” The World-View of Prehistoric Man: Papers Presented at a Symposium in Lund, 5-7 May 1997. Pp. 93-102. Stockholm, 1998, p. 100). They occur in small demes practicing the rule of bilateral cross-cousin marriage that repeats from generation to generation and creates a special kin-terminological signature in the form of identical consanguineal and affinal terms. (For Karitiana as having a Dravidian kin terminology, albeit with lots of unique features, see Landin, Rachel M. Kinship and Naming among the Karitiana of Northwestern Brazil. M.A. thesis. University of Texas – Austin, 1989). Once population size grows, these logically most elementary systems open up allowing for marriage between more distant or entirely unrelated people. Correspondingly, kin terminologies lose the consanguineal-affinal equations. Interestingly enough, Maya show a completely different genetic pattern, the one with moderate IBD values (see table above). However, among the Lacandons, anthropologists (see, e.g., Boremanse, Didier. A Comparative Study of Two Maya Kinship Systems. Sociologus 31 (1981): 1-37) described a kinship systems reminiscent of an ancient “symmetric-prescriptive” prototype. It’s likely that the samples at geneticists’ disposal didn’t come from the isolated Lacandon communities but rather from more cosmopolitan Mayan populations. In the past, geneticists (see, e.g., Merriwether, Reed and Ferrell. “Ancient and contemporary mitochondrial DNA variation in the Maya,” Bones of the Maya : studies of ancient skeletons. Washington, 1997) reported high levels of genetic diversity among Maya that were matched in the Americas only by such exceptionally diverse populations as Nuu-chah-nulth. Admixture, including recent admixture with Europeans, should be able to explain this surge in diversity and decline of IBD.