Molecular Variability of the 16p13.3 Region in American Indians
Annals of Human Genetics (2007) 71, 64–76 DOI: 10.1111/j.1469-1809.2006.00296.x
Molecular Variability of the 16p13.3 Region in Amerindians and Its Anthropological Significance
Battilana, J., L. Cardoso-Silva, R. Barrantes, K. Hill, A. M. Hurtado, F. M. Salzano and S. L. Bonatto
A total of 1558 base pairs in the 16p13.3 region were investigated in 98 individuals of Mongolian, Northern Arctic and Amerindian affiliation, and the results compared with those obtained in a previous worldwide study of the same genomic region. Fifty-five polymorphic sites could be classified into thirty-five haplotypes from the total data. A median joining network based on the haplotypes revealed two distinct clusters: one with low diversity, with haplotypes found in all five geographic-ethnic categories; while the other, with the most divergent haplotypes, was composed mainly of Africans and a few Amerindians. Almost all neutrality parameters yielded significantly negative values. Demographic simulations with the exclusively Amerindian dataset rejected all scenarios, including a bottleneck beginning more than 12,000 years ago. The demographic scenarios tested considering population growth were similar among the Amerindian and worldwide or Eurasian data sets. The results suggest that Amerindians are a representative sample of Eurasian populations, preserving the signal of demographic growth from the out of Africa exodus and, together with data from uniparental markers, support a scenario of a bottleneck of moderate intensity during the peopling of the New World.
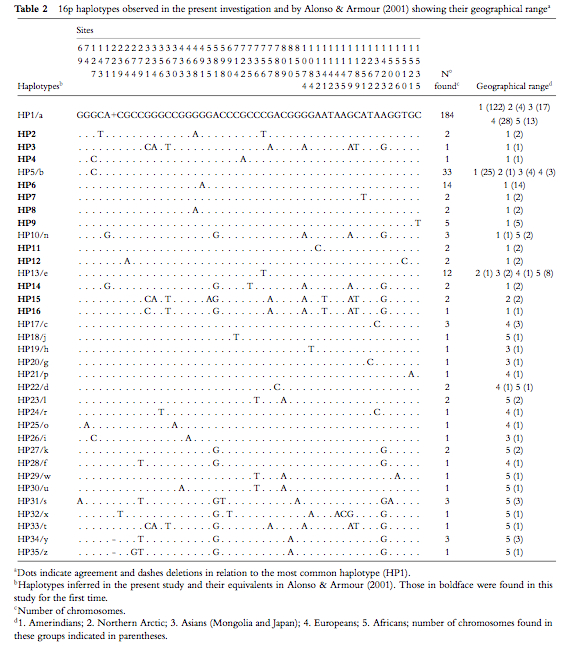
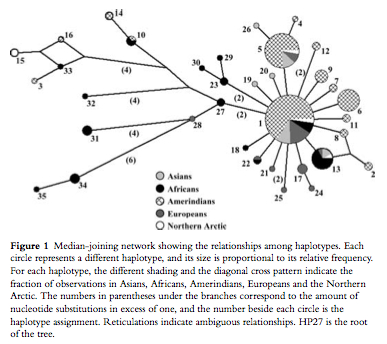
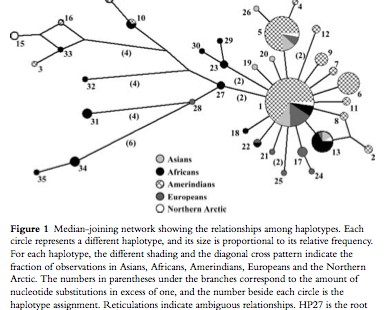
This somewhat oldish paper provides haplotype definitions and a phylogenetic tree for a highly polymorphic autosomal region assumed to be free of genetic hitchhiking and governed by neutral mutation dynamics by virtue of being shielded behind another region with a high recombination rate. Battilana et al. (2007) build on to the pioneering effort by Santos Alonso & John Armour (“A highly variable segment of human subterminal 16p reveals a history of population growth for modern humans outside Africa,” Proc Natl Acad Sci USA 98 (2001), 864-869) to analyze this interesting genomic locus by adding an American Indian sample to Alonso & Armour Old World sample. The Battilana et al. (2007) results were surprising: instead of the expected peripheral position of American Indians in a global haplotype phylogeny, they obtained a global median-joining network centered on the most frequent (62% of the chromosomes) haplotype (haplotype 1, Table 2 below) found on every continent but achieving its highest frequencies in American Indians (Fig. 1 below). (Alonso & Armour postulated haplotype 27 as the root of the tree, but they didn’t have an American Indian sample.)
The median-joining network is effectively split into two parts: the star-like cluster on the right (Fig. 1 above) is squarely centered on haplotype 1 surrounded by a bunch of one- and two-step derivations most of which are geographically restricted to American Indian, European, Asian and African populations; a set of highly divergent (4-to-6 steps away from the nodal haplotype 1) haplotypes on the left includes mostly African-specific haplotypes but also American Indian, Chukchee and Basque haplotypes. Although Batillana et al. (2007) didn’t propose an out-of-America interpretation of their haplotype network (but they alluded to something of that sort in their phrase “a strong population expansion from a restricted source,” p. 73), they did reject the recent model for the peopling of the Americas on the basis of the fact that American Indians look not like a population that underwent a bottleneck but rather like a fully representative sample of Old World variation participating in the same population growth spurt present in Old World populations. (As a side note, this is consistent with Victor Grauer’s musicological argument suggesting that the New World must have been peopled during the very same first wave of out-of-Africa colonization because South American polyphonic musical traditions are unmistakably similar to those found in Pygmies, Khoisans and some Papua New Guinea groups.)
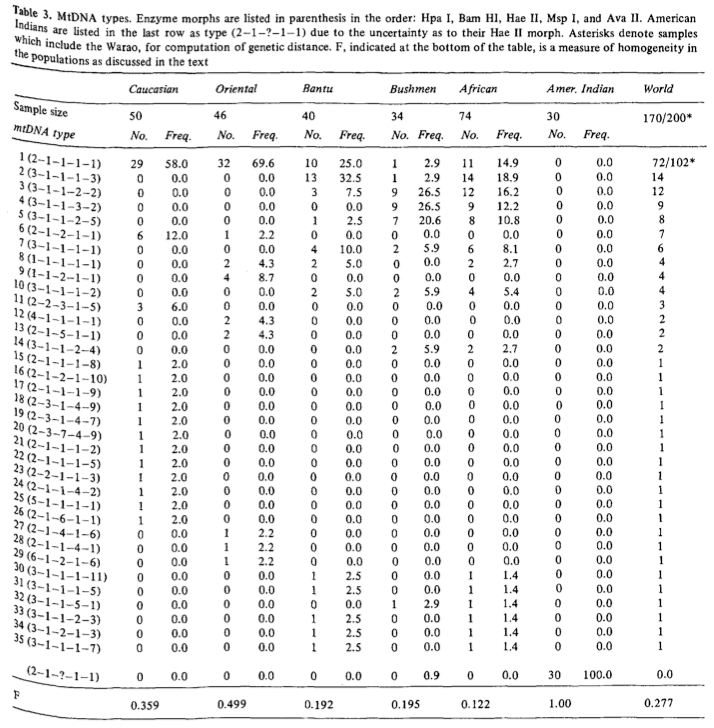
What is remarkable is the uncanny resemblance between Batillana et al.’s Table 2 and Fig. 1 and Table 3 and Fig. 6 (see below) from the first worldwide study of mtDNA variation by Johnson et al. (“Radiation of Human Mitochondria DNA Types Analyze by Restriction Endonuclease Cleavage Patterns,” J Mol Evol 19 (1983): 255-271).
In a manner similar to haplotype 1 in Batillana et al. (2007), the nodal mtDNA haplotype 1 was found at highest frequencies in the New World and it progressively declined as one moved from America to Asia to Europe and, finally, to Africa. The overall impression one might get from both Johnson et al (1983) and Batillana et al. (2007) is that a population expansion originating in the Americas resulted in worldwide dissemination of a nodal haplotype with a) the corresponding emergence of derived haplotypes at a varying mutation rate, most aggressively in Africa but also in European Basques, Circumarctic populations and some Amerindian groups, and b) with the conservation of the nodal haplotype in the Americas due to geographic isolation.