Neandertal Admixture in microRNA Genes
Molecular Biology and Evolution (27 January 2012), doi:10.1093/molbev.
An Ancestral miR-1304 Allele Present in Neanderthals Regulates Genes Involved in Enamel Formation and Could Explain Dental Differences with Modern Humans
Lopez-Valenzuela, Maria, Oscar Ramirez, Antonio Rosas, Samuel Garcıa-Vargas, Marco de la Rasilla, Carles Lalueza-Fox, and Yolanda Espinosa-Parrilla
Genetic changes in regulatory elements are likely to result in phenotypic effects that might explain population-specific as well as species-specific traits. MicroRNAs (miRNAs) are posttranscriptional repressors involved in the control of almost every biological process. These small noncoding RNAs are present in various phylogenetic groups, and a large number of them remain highly conserved at the sequence level. MicroRNA-mediated regulation depends on perfect matching between the seven nucleotides of its seed region and the target sequence usually located at the 3# untranslated region of the regulated gene. Hence, even single changes in seed regions are predicted to be deleterious as they may affect miRNA target specificity. In accordance to this, purifying selection has strongly acted on these regions. Comparison between the genomes of present-day humans from various populations, Neanderthal, and other nonhuman primates showed an miRNA, miR-1304, that carries a polymorphism on its seed region. The ancestral allele is found in Neanderthal, nonhuman primates, at low frequency (5%) in modern Asian populations and rarely in Africans. Using miRNA target site prediction algorithms, we found that the derived allele increases the number of putative target genes for the derived miRNA more than ten-fold, indicating an important functional evolution for miR-1304. Analysis of the predicted targets for derived miR- 1304 indicates an association with behavior and nervous system development and function. Two of the predicted target genes for the ancestral miR-1304 allele are important genes for teeth formation, enamelin, and amelotin. MicroRNA overexpression experiments using a luciferase-based assay showed that the ancestral version of miR-1304 reduces the enamelin- and amelotin-associated reporter gene expression by 50%, whereas the derived miR-1304 does not have any effect. Deletion of the corresponding target sites for miR-1304 in these dental genes avoided their repression, which further supports their regulation by the ancestral miR-1304. Morphological studies described several differences in the dentition of Neanderthals and present-day humans like slower dentition timing and thicker enamel for present-day humans. The observed miR-1304-mediated regulation of enamelin and amelotin could at least partially underlie these differences between the two Homo species as well as other still-unraveled phenotypic differences among modern human populations.
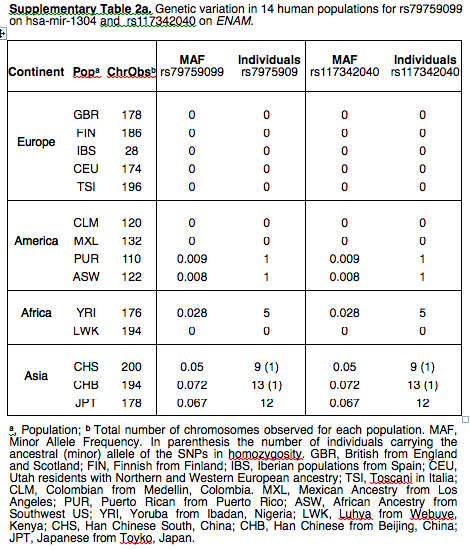
Every paper claiming “admixture” with archaic hominins, especially Neandertals, is important. This one reports on a unusual pattern of “Neandertal” introgression. The ancestral allele (rs79759099) was found in Neandertals, nonhuman primates, at low frequency (5%) in modern Asian populations and only in one African from Kenya. It was not detected in Europeans, East Africans, and, as Supplemental Materials indicate, in American Indians (see below). Minimal percentages recorded in Puerto Rico and Southwest US come from African Americans, which means that the allele is attested in West Africa beyond the one Kenyan individual.
The most interesting part of the paper didn’t make it into the abstract. The authors struggle to explain the unusual pattern of distribution of the ancestral allele and they consider several possibilities:
“Interestingly, analysis of 1000 Genomes Project data revealed that this nucleotide change is not fixed in present-day human populations, and it turned out to be an SNP (rs79759099) present in Asian populations (allele frequency of about 0.06). In European and East-African populations, the ancestral allele was not found, and populations with West-African origin carry the ancestral allele but at very low frequencies. Since this distribution schema could be the result of selective sweeps within recent human populations compatible with a beneficial role for the new derived miR-1304 allele, we looked for signatures of selection along the region using HapMap and HGDP data, but no evidences for a selective sweep, such as an excess rare allele variants or a block of extensive linkage disequilibrium, could be found. Other explanation for the presence of the ancestral allele in Asian populations could be that it was reintroduced through admixture with Neanderthals; however, because Neanderthal admixture is restricted to non-African populations (Green et al. 2010) and given the presence of the ancestral miR- 1304 in some Africans and its low frequency in Asia, the distribution of the ancestral miR-1304 could not solely be explained in terms of introgression. Furthermore, miR-1304 is not included into the 13 regions that have been identified as candidate gene flow regions between Neanderthals and modern humans that were described through the analysis on extended Neanderthal haplotype blocks in modern human genomes (Green et al. 2010). In this scenario, a combination of admixture in Asia such as the described by Skoglund (Skoglund and Jakobsson 2011) with rs79759099 being a retained ancient polymorphism in Africa could be the most plausible explanation.”
After dismissing the popular “admixture in non-Africans” theory, their “most plausible explanation” is essentially retention in West Africa and introgression in Asia. This interpretation, however, faces its own challenges. If the allele introgressed into modern humans in Asia from Neandertals, why didn’t it happen in Europe where most Neandertals lived. (Curiously, the authors don’t seem to know that Neandertal fossils and tools were found in northeast Asia, although this would fit their data, as they declare (p. 2) that Neandertals “inhabited parts of Europe and Western Asia”). If introgression occurred in West Asia, why wasn’t Europe affected, especially since Upper Paleolithic technologies are shared between North Africa, West Asia and Europe. If introgression occurred in northeast Asia (e.g., in South Siberia where Neandertal and Denisovan fossils were found), as the presence of the ancestral allele in Chinese and Japanese suggests, then why wasn’t it detected in the New World, which was likely colonized from that area? If Africa is the continent on which the ancestral allele survived, then why its present-day frequencies in Africa have such a low frequency and restricted distribution?
An even more plausible explanation is that introgression occurred in Asia (and under Out-of-America II American Indians wouldn’t be affected) and then the admixed population migrated to West Africa (the distribution of Y-DNA hg DE connecting Asia and Africa with the exclusion of America and Europe comes to mind as a possible analogy), whereas the original unadmixed population migrated to Europe.
Needless to say, this scenario is only good with the data at hand. It’s always possible that better sampling on all continents will alter the picture and inspire a whole another interpretation.