Genetics and Linguistics of the Bantu Expansion
Proc. Roy. Soc. Lond. B 279: 3256-3263. doi: 10.1098/rspb.2012.0318
Bringing Together Linguistic and Genetic Evidence to Test the Bantu Expansion
De Filippo, Cesare, Koen Bostoen, Mark Stoneking, and Brigitte Pakendorf.
The expansion of Bantu languages represents one of the most momentous events in the history of Africa. While it is well accepted that Bantu languages spread from their homeland (Cameroon/Nigeria) approximately 5000 years ago (ya), there is no consensus about the timing and geographical routes underlying this expansion. Two main models of Bantu expansion have been suggested: The ‘early-split’ model claims that the most recent ancestor of Eastern languages expanded north of the rainforest towards the Great Lakes region approximately 4000 ya, while the ‘late-split’ model proposes that Eastern languages diversified from Western languages south of the rainforest approximately 2000 ya. Furthermore, it is unclear whether the language dispersal was coupled with the movement of people, raising the question of language shift versus demic diffusion. We use a novel approach taking into account both the spatial and temporal predictions of the two models and formally test these predictions with linguistic and genetic data. Our results show evidence for a demic diffusion in the genetic data, which is confirmed by the correlations between genetic and linguistic distances. While there is little support for the early-split model, the late-split model shows a relatively good fit to the data. Our analyses demonstrate that subsequent contact among languages/populations strongly affected the signal of the initial migration via isolation by distance.
A team of Max-Planck geneticists with the help of linguists Koen Bostoen and Brigitte Pakendorf, published another paper on the Bantu expansion. In my earlier post, I quoted the 2011 paper from a broader team (De Filippo et al. (2011). “Y-Chromosomal Variation in Sub-Saharan Africa: Insights Into the History of Niger-Congo Groups,” Molecular Biology and Evolution 28 (3): 1255-1269) and reported on the recent partnership between Bostoen and anthropological linguist Jeff Marck on the reconstruction of Bantu kinship and social organization.
De Filippo et al. (2012) seem to have arrived at very definitive solution to the problem of the timing and demography of the Bantu expansion. They argue that a) there’s close parallelism between the linguistic kinship between Bantu languages and the genetic kinship between their speakers; b) the Bantu expansion was a demic diffusion; c) it began around 5,000 BP; d) East Bantu were a later split from West Bantu and not an independent migration from West Africa; d) subsequent genetic and linguistic contacts blurred the original migration signal.
I can only applaud to the growing partnership between geneticists and linguists and the gradual incorporation of kinship data into broader syntheses. And the close correspondence between genetic and linguistic distances in the Bantu area confirms my overall impression that genes and languages correlate well. This is especially true for recently expanded families. As time goes by, this correspondence, however, weakens due to varying genetic contacts between the now-isolated branches of the family. Time tends to complicate the search for both linguistic signs of kinship and the correspondences between languages and genes.
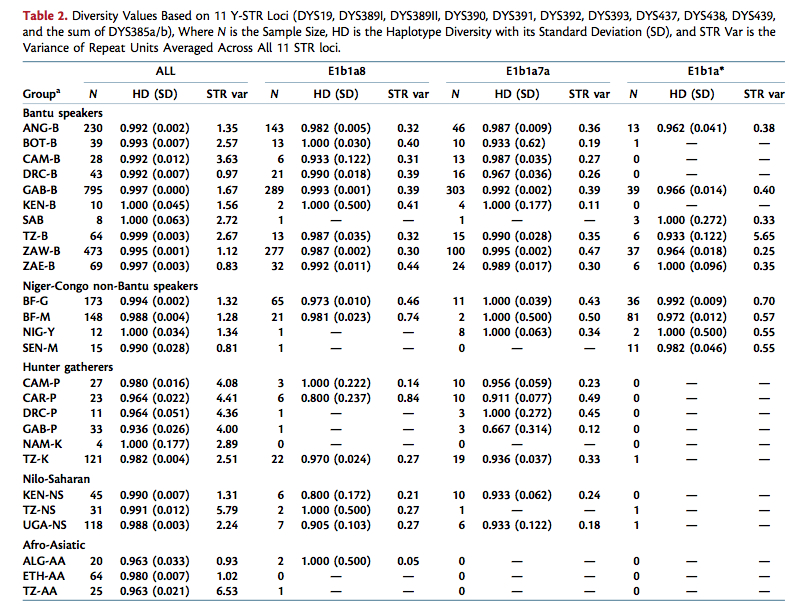
Still, there some contradictions between De Filippo (2011) and De Filippo (2012) as well as ambiguities in the genetic data that will undoubtedly lead to discussion and further research. De Filippo (2011) couldn’t detect a steady trend of decreasing Y-DNA diversity with increasing distance from the Bantu homeland in West Africa (see their Table 2 below). In fact, haplotype diversity (HD) values were higher among Bantu-speakers than among Niger-Congo non-Bantu-speakers (Mande, Gur, etc.) located close to the Bantu homeland in West Africa.
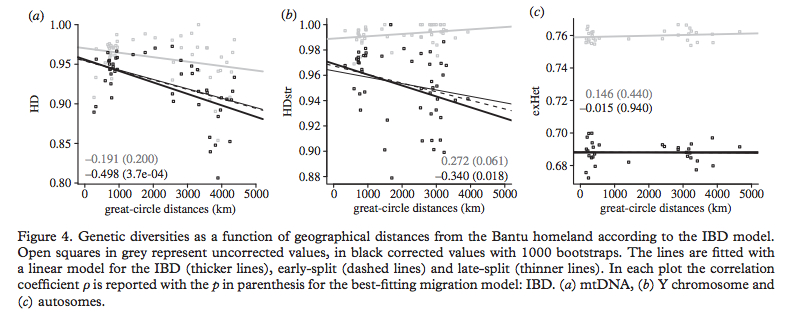
De Filippo (2012, 4) corrected for sample size and reported the much needed finding that “mtDNA and Y- chromosomal haplotype diversity decreased significantly with increasing distance from the Bantu homeland.” The heterozygosity of the autosomes, on the other hand, remained flat (see below, Fig. 4).
How robust is then the inference of genetic diversity decay with increasing distance from the homeland? The authors undermine the confidence that it is by saying:
“given that mtDNA and the Y chromosome are basically single loci with different mutation rates it is difficult to disentangle the effects of stochasticity from demographic events. In addition, the sparse sampling of Bantu populations in general, and the mismatch between the availability of mtDNA versus Y-chromosomal data in particular, precludes any definitive conclusions about the nature of the Bantu migrations.” (p. 6).
This means that with additional sampling the correlation, already unsteady at this stage, may change unpredictably.
De Filippo et al. (2012) detected a much stronger fit between a late-split model of Bantu subgrouping and genetic distances than between an early-split model and genetic distances. It means that the Bantu expansion went first straight down south across the equatorial forest and then there was a further migration from these newly colonized areas eastward.
Most interestingly for a student of kinship is the authors’ finding that mtDNA haplotype diversity decreased with distance from homeland much more than Y-DNA haplotype diversity (see, again, their Fig. 4 above). This contradicts their 2011 finding that patrilocality and polygyny must have reduced Y-DNA diversity in Bantu. While the new finding jibes well with Marck & Bostoen’s reconstruction of matrilocality for the expanding Bantu populations with later, multiple convergent shifts to patrilocality, the different genetic signals of reduced mtDNA vs. reduced Y-DNA diversity still need to be teased out and stratified historically.