mtDNA and Y-DNA Markers in South American Indians
Genetics and Molecular Biology 2012 Apr-Jun; 35 (2): 365-387. doi: 10.1590/S1415-47572012005000027.
Uniparental Genetic Markers in South Amerindians
Rafael Bisso-Machado, Maria C. Bortolini, and Francisco M. Salzano.
A comprehensive review of uniparental systems in South Amerindians was undertaken. Variability in the Y-chromosome haplogroups were assessed in 68 populations and 1,814 individuals whereas that of Y-STR markers was assessed in 29 populations and 590 subjects. Variability in the mitochondrial DNA (mtDNA) haplogroup was examined in 108 populations and 6,697 persons, and sequencing studies used either the complete mtDNA genome or the highly variable segments 1 and 2. The diversity of the markers made it difficult to establish a general picture of Y-chromosome variability in the populations studied. However, haplogroup Q1a3a* was almost always the most prevalent whereas Q1a3* occurred equally in all regions, which suggested its prevalence among the early colonizers. The STR allele frequencies were used to derive a possible ancient Native American Q-clade chromosome haplotype and five of six STR loci showed significant geographic variation. Geographic and linguistic factors moderately influenced the mtDNA distributions (6% and 7%, respectively) and mtDNA haplogroups A and D correlated positively and negatively, respectively, with latitude. The data analyzed here provide rich material for understanding the biological history of South Amerindians and can serve as a basis for comparative studies involving other types of data, such as cultural data.
Link (Free Access PDF)
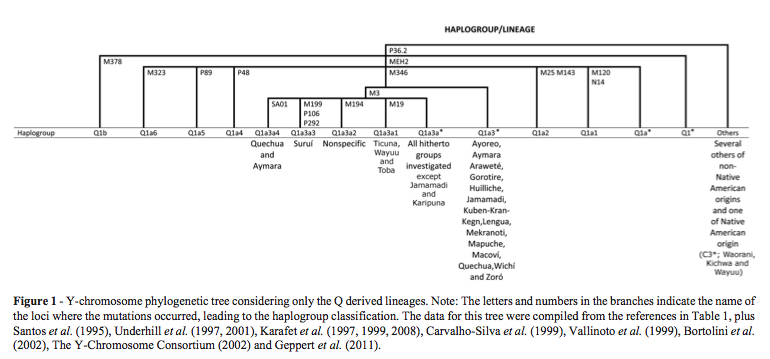
A very comprehensive summary of the distribution of mtDNA and Y-DNA markers in South American Indian populations across linguistic families and geographics. Two callouts:
1. All four principal American Indian mtDNA haplogroups (A, B, C, D) are found in nearly all linguistic families and geographic regions in South America. This is consistent with ancient DNA studies.
2. Y-DNA hg C* is present in Northwest South Amerindian populations, per Geppert et al. 2011. (“Hierarchical Y-SNP assay to study the hidden diversity and phylogenetic relationship of native populations in South America,” Forensic Sci Int Genetics 5 (2):100-104). This finding dispels the notion that hg C3, so far mostly attested in North American Indians and Inuits as C3b, is the result of a separate, later migration from Asia. Although neither Geppert et al. (2011), nor Bisso-Machado et al. (2012) report all the SNPs for the South American Indian C*, it’s likely that it’s closer to North American Indian C3b than to any Old World Cs.
Just like mtDNA, Y-DNA shows that American Indians are inherently polymorphic and this diversity is attested in both North and South. At the same time, one cannot help but notice that the American Indian Y-DNA picture is heavily dominated by Q lineages, whereas the mtDNA picture is more balanced between macrohaplogroup M (C, D) and macrohaplogroup N (A, B) lineages. This could indicate the long-term prevalence of patrilocality in South America, which drives Y-DNA diversity down, but ethnographically this is far from being always the case, with uxorilocality being the prevalent form of post-marital residence in, for example, Amazonia.