mtDNA CR HVS1 16189
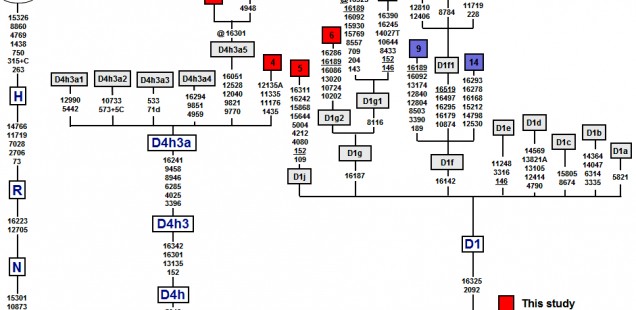
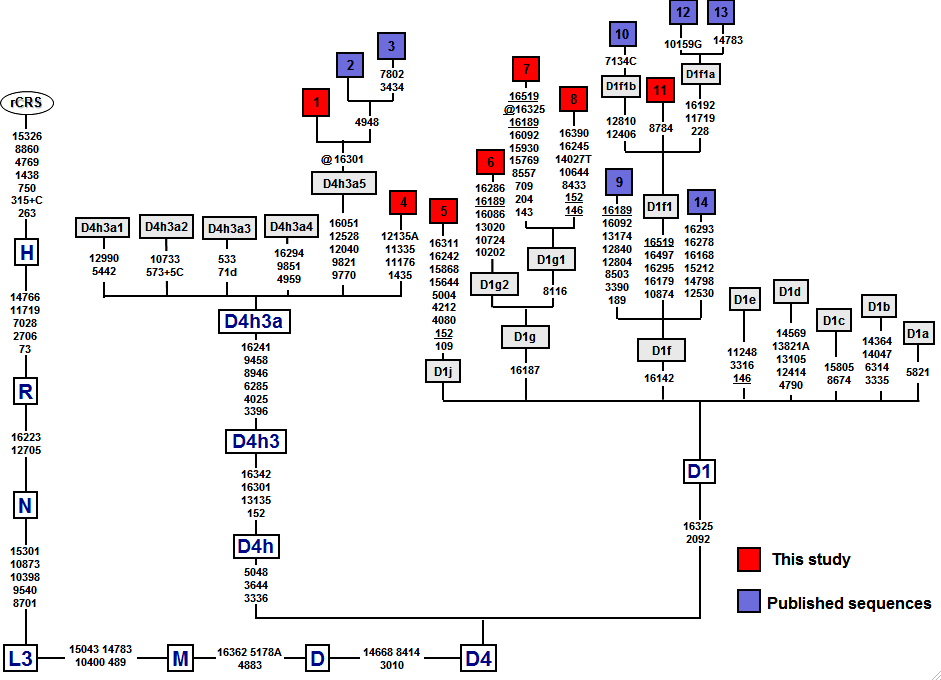
In the new paper on mtDNA lineages in the populations of Patagonia and Tierra del Fuego, de Saint Pierre et al. (2012) published several new D1 sequences from this region of South America. Among them D1g2 and D1g1 are interesting because they are both marked by mutations back to ancestral state at np 16189 (see below).
Neandertals attest to a C at this position. mtDNA phylogeneticists postulate that at the African lineage ancestral to L2, L3, L4 and L6 haplogroups transition C16189T occurred (see below, from PhyloTree).
So for hundreds of thousands of years since the Neandertal-modern human split and for tens of thousands of years since the purported Khoisan-modern human split, this site did not mutate. But then an avalanche of back mutations at 16189 affected a large number of derived lineages, cutting across haplogroup L2, macrohaplogroups M, N and R. In Africa, L2a1b, L2a1i, L2a2’3’4, L2a5, L2d, L4b1, L3b2a, L3d3a, L3e2b and M1 have the back mutation. In West Eurasia, T1, T2f, R0a1b, HV1b2, H1a6, H1b, H1c3b, H1ap1, H3av, U1a’c, U5a1d2a, U5b1a, U5b2a, U5b2a1a (double back mutation), U5b2b4, U6a1a, U6a2, U6c, U2d, U2e, U4d2, U8b1, K1a2a1, K1b1a1b, K1e1, K2a8, N1a1a1a, I1a1d. In Asia, M2b, M3c1, M38b, M37e, M45, M5a3b, M7a1a2, M7b2, M8a2a, M13a, M53, M75, M41a, M27a, M29, M35c, M42b, M47, M76, M77, M80, C4c1b, C7a2, Z4a, G2a1c, G2a1d2 (double back mutation), G3a2, D4b2a2a, D4b2b2c, D4e4a1, D5, F1b, R11’B6, R30b1, R31b, N3, N9a5, N9b, Y1a, N10a, N11a1, A4b, A7, X, X2e1. In America C1b4, D1a1, D1g2, D1g1, A2e. This site is considered hypervariable but it is unclear why there is a need to postulate the original C16189T transition at the root of L2’L3’L4’L6 instead of treating all the “back mutations” to the ancestral state found everywhere around the globe, on all possible lineages as retentions from the Neandertal-modern human nexus. Just like L0 and L1 lineages are.
Earlier, I asked a similar question about 9bp deletion, which displays similar behavior to C16189T, is also found on closely related lineages (e.g., B4’B5 and R11’B6) and, incidentally, occurs in tandem with ancestral C16189 in Africa, West Eurasia, Southeast Asia and America. While existing Neandertal sequences do not have 9bp deletion, there are too few archaic sequences available to ascertain that archaic hominins were monomorphic here.



This is a very intriguing subject for me. I’ve been trying to figure out the whole Neanderthal phenomenon ever since I began researching my own mtDNA. My own haplotype (now tentatively classified “W1e”; originally called just “X”) is unlike all of my near relations with the same classification. Of 13 closely related near-matches (in the mitosearch databank), mine is the only one with six mutations (the rest have fewer). Mine also has the 16189C mutation, but I don’t know whether it’s a retention or a “back mutation”. I wonder how that mutation came to be associated with Neanderthal hybridization? Is it because it’s also found in known Neanderthal DNA? Or, does it correlate with the B006 or some other “Neanderthal” marker? I get the impression that geneticists might possibly have the whole Neanderthal definition backwards or upside-down, somehow. Could it be that some of the markers attributed to the Neanderthal, are in fact ~Human (Sapiens) aspects of the same creature? After all, the genetic definition of “Neanderthal” has been formed around preconceived theories about its origins. Many geneticists today, speak of the Neanderthal as an “archaic” ~ancestor of modern Humans, which in a way it is (but only for ~some of us). However, John Pfeiffer in “The Emergence of Man” and other early Anthropologists (prior to modern DNA analysis) described the Neanderthal as a later hybrid ~subspecies of Homo Sapiens. And I’ve seen no real evidence, genetic or otherwise, to believe that it isn’t just that. In fact, scientists seem to be dancing all around the subject of the Neanderthal, never stating any solid conclusions about it. (Of course, the Denisovan and other little understood hominids just adds more complexity to the whole puzzle).
German, I would like to re-post this quote here, if correct, it supports the out of America I and a New World primate link for our species from An America Wellspring, for the record. Please email me with verification of posting, thank you very much for your Blog and your open discourse.
“The apparent absence of Denisova ancestry in Native Americans in our study could be influenced by the biased affinity to the Neandertal genome that is expected because of ascertainment bias and genetic drift, but analyses of unascertained low-coverage shotgun sequence data from a single Native American individual resulted in a similar conclusion (24). If absent in America, the Denisova component must have appeared in the ancestors of East Asians after their divergence from Native Americans, which has been dated to ∼14–30 kya (58, 60, 61) (Skoglund and Jakobsson 2011). They reference ((24) .. 24.; Reich D, et al. (2010) Genetic history of an archaic hominin group from Denisova Cave. (from Nature 468:1053–1060 2011).
Native Americans, our ancestors, did not diverge from East Asians nor do we need to trace our-self back to Old World hominid lineages.