mtDNA from the South Cone of South America
PLoS ONE 7(9): e43486. doi:10.1371/journal.pone.0043486
An Alternative Model for the Early Peopling of Southern South America Revealed by Analyses of Three Mitochondrial DNA Haplogroups
Michelle de Saint Pierre, Claudio M. Bravi, Josefina M. B. Motti, Noriyuki Fuku, Masashi Tanaka, Elena Llop, Sandro L. Bonatto, Mauricio Moraga
After several years of research, there is now a consensus that America was populated from Asia through Beringia, probably at the end of the Pleistocene. But many details such as the timing, route(s), and origin of the first settlers remain uncertain. In the last decade genetic evidence has taken on a major role in elucidating the peopling of the Americas. To study the early peopling of South America, we sequenced the control region of mitochondrial DNA from 300 individuals belonging to indigenous populations of Chile and Argentina, and also obtained seven complete mitochondrial DNA sequences. We identified two novel mtDNA monophyletic clades, preliminarily designated B2l and C1b13, which together with the recently described D1g sub-haplogroup have locally high frequencies and are basically restricted to populations from the extreme south of South America. The estimated ages of D1g and B2l, about ~15,000 years BP, together with their similar population dynamics and the high haplotype diversity shown by the networks, suggests that they probably appeared soon after the arrival of the first settlers and agrees with the dating of the earliest archaeological sites in South America (Monte Verde, Chile, 14,500 BP). One further sub-haplogroup, D4h3a5, appears to be restricted to Fuegian-Patagonian populations and reinforces our hypothesis of the continuity of the current Patagonian populations with the initial founders. Our results indicate that the extant native populations inhabiting South Chile and Argentina are a group which had a common origin, and suggest a population break between the extreme south of South America and the more northern part of the continent. Thus the early colonization process was not just an expansion from north to south, but also included movements across the Andes.
Link (Free Full Text PDF)
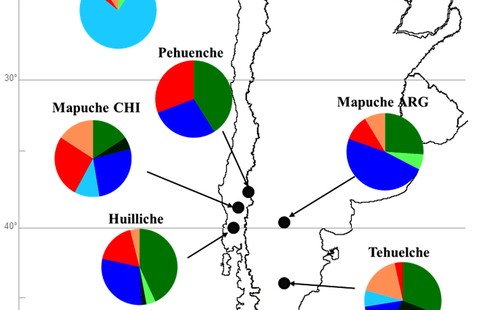
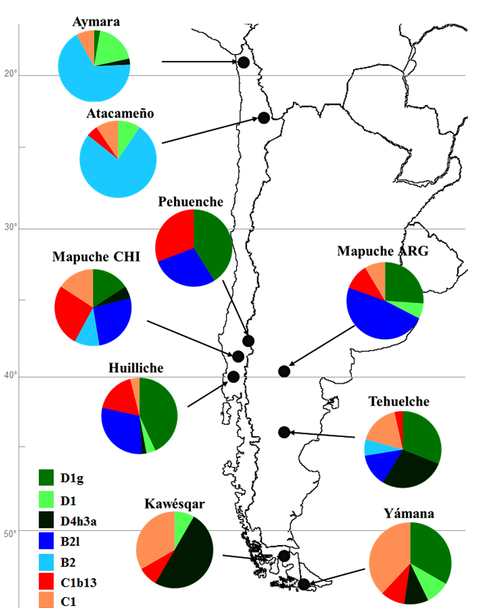
This is a comprehensive mtDNA study of the most remote New World region, which suffered massive native depopulation as a result of European colonization. The authors depict it as a refugium that immediately attracted the earliest colonists in South America who took the coastal route from Alaska to Monte Verde, then across the Andes to Tierra del Fuego, and then remained largely sealed away from subsequent gene flow from the north until the arrival of the Spaniards. The authors describe two novel mtDNA lineages – B2l and C1b13 – and propose changes in lineage definition for hgs D1g and D4h3a5. The discovery of novel lineages is ongoing in the New World, but caution needs to be exercised when these new lineages are from clades wide-spread in Asia, as these can be products of recent admixture with Asian immigrants to the Americas. In this case, the novel lineages belong to established American Indian clades, so this concern does not apply. The distribution of all the detected haplotypes is below.
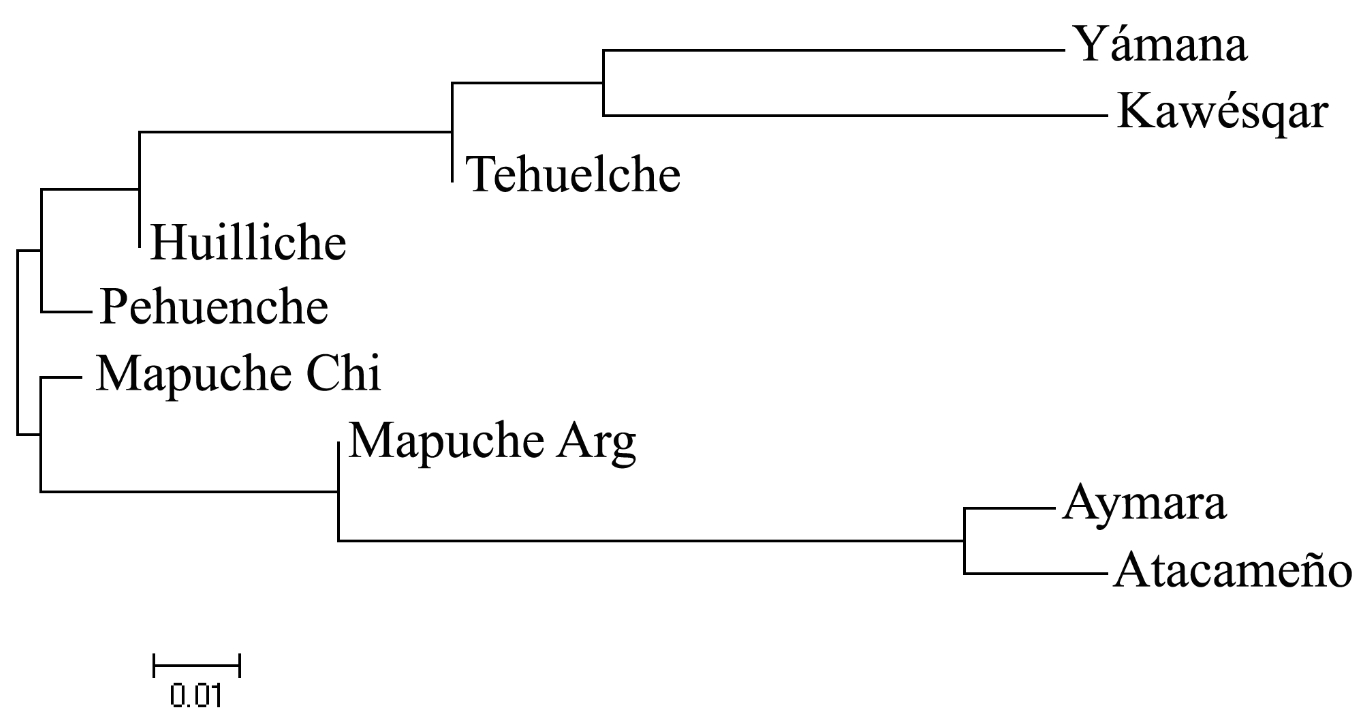
A dendrogram built on the basis of Fst differences between the populations shows their mutual relationships.
Mapuche presumably stayed behind on the coastal side of the Andes, while Kaweskar and Yamana formed their own clade by settling in Tierra del Fuego. This region virtually lacks hg A2, which is very common in northern North America and around Beringia. Hg B2 is attested everywhere but the isolated Tierra del Fuego cluster. One can hypothesize that Yamana and Kaweskar used to have it but they lost it either through drift prior to European colonization or in the course of the recent depopulation caused by the European invasion. The same concerns hg D1g, which is very common in south-central Chile and Argentina and is found in ancient remains in Tierra del Fuego but was not detected in Kaweskar. All Patagonian and Tierra del Fuego populations have hg D4h3a5. The localized presence of the lineages is interpreted by de Saint Pierre et al. (2012) as evidence for genetic continuity between contemporary populations and the founders. (This has bearing on the argument advanced by paleobiologists for craniological discontinuity between Paleoindian and modern American Indians suggestive of two migration waves to the New World.) Hg D4h3a tends to have a coastal distribution in the New World. In North America it is limited to southern California (16% Chumash) but is attested in ancient remains from the Klunk Mound (Illinois) and the On Your Knees Cave (SE Alaska). Still, the question looms large as to why hg D4h3a is so deeply rooted in South America, while being nearly completely absent in northern North America. One sequence assigned to the sister clade D4h3b was detected in China (Perego, et al. 2009. “Distinctive Paleo-Indian Migration Routes from Beringia Marked by Two Rare mtDNA Haplogroups,” Curr Biol 19, 2) continuing the pattern seen in northern North America.
The regional gaps in the distribution of mtDNA haplotypes in the New World (no hg B and hg D4h3 in northern North America, no hg B in Tierra del Fuego, no hg A2 in Patagonia and Tierra del Fuego, no D1g in Kaweskar, etc.) seem to have come about in the last 10,000 years, as in virtually every case ancient remains confirm their presence in that region).
De Saint Pierre et al. (2012) are willing to grant some extra antiquity to the peopling of the South Cone of South America:
“For a specific lineage restricted to the end of the Southern Cone of South America, the dates for D1g and B2l are old and allow us to support a scenario for the settlement of America with dates of the first entry between 18-20 Kyr, the first settlers having taken ~3–5 Kyr to cross the continents and reach Patagonia.”
They note that intragroup genetic diversity metrics in Patagonia and Tierra del Fuego are high, with haplotype diversity (Hd) reaching 1 in Aymara and Atacameño. The full spectrum of diversity metrics can be seen below.

n: sample.
h: number of haplotypes.
S: number of polymorphic sites.
Hd: haplotype diversity.
K: mean number of pairwise differences.
π: nucleotide diversity.
Now let’s compare them with the metrics obtained from mtDNA sequences in South Siberia, an enclave in Northeast Asia from which many hypothesize the colonization of the Americas began (see below, from Derenko et al. 2003. “Diversity of Mitochondrial DNA Lineages in South Siberia,” Ann Hum Gen 67, p. 406):
 It is noteworthy that the metrics (organized by sequence in De Saint Pierre et al. and by population by Derenko et al.) are very similar between the two regions: haplotype diversity and number of lineages are higher in the South Cone of South America, mean number of pairwise differences is about the same, nucleotide diversity is higher in South Siberia. Assuming that the settlers of the South Cone had to traverse all of the Americas in 3-4,000 years (as De Saint Pierre et al. suggest), which would have put them under a demographic pressure, there does not seem to be much evidence for a bottleneck out of Asia.
It is noteworthy that the metrics (organized by sequence in De Saint Pierre et al. and by population by Derenko et al.) are very similar between the two regions: haplotype diversity and number of lineages are higher in the South Cone of South America, mean number of pairwise differences is about the same, nucleotide diversity is higher in South Siberia. Assuming that the settlers of the South Cone had to traverse all of the Americas in 3-4,000 years (as De Saint Pierre et al. suggest), which would have put them under a demographic pressure, there does not seem to be much evidence for a bottleneck out of Asia.
Where the differences between Asia and America are sharp is in the Fst metrics that measure intergroup diversity.
 The highest value in Asia is the Uyghur-Tofalar Fst of 0.193, the highest value in the South Cone is 0.294 between Atacameno and Yamana. High Fst is a trademark of American Indian population structure (with an attested antecedent in Denisovans) and it indicates high levels of population fragmentation in the New World.
The highest value in Asia is the Uyghur-Tofalar Fst of 0.193, the highest value in the South Cone is 0.294 between Atacameno and Yamana. High Fst is a trademark of American Indian population structure (with an attested antecedent in Denisovans) and it indicates high levels of population fragmentation in the New World.



Am I wrong to assume:
1. There could be 2 sources of Denisovian DNA sequences in the lower cone of South America (after 5000-7000 y BCA?
2. Is the attested original 20K BCA DNA from Tierra Dela Fuego more like Melanesian and Australian Aboriginal DNA?
3. Do the Denisovian contribution from the documented Beringial populations (that apparently pushed the darker Melanonesian immigrant southward) exceed that of the Melanesian contributions?
Thanks.