A High Coverage of the Denisovan Hominin
Science 30 August 2012 DOI: 10.1126/science.1224344
A High-Coverage Genome Sequence from an Archaic Denisovan Individual
Matthias Meyer, Martin Kircher, Marie-Theres Gansauge, Heng Li, Fernando Racimo, Swapan Mallick, Joshua G. Schraiber, Flora Jay, Kay Prüfer, Cesare de Filippo, Peter H. Sudmant, Can Alkan, Qiaomei Fu, Ron Do, Nadin Rohland, Arti Tandon, Michael Siebauer, Richard E. Green, Katarzyna Bryc, Adrian W. Briggs, Udo Stenzel, Jesse Dabney, Jay Shendure, Jacob Kitzman, Michael F. Hammer, Michael V. Shunkov, Anatoli P. Derevianko, Nick Patterson, Aida M. Andrés, Evan E. Eichler, Montgomery Slatkin, David Reich, Janet Kelso, Svante Pääbo.
We present a DNA library preparation method that has allowed us to reconstruct a high-coverage (30X) genome sequence of a Denisovan, an extinct relative of Neandertals. The quality of this genome allows a direct estimation of Denisovan heterozygosity, indicating that genetic diversity in these archaic hominins was extremely low. It also allows tentative dating of the specimen on the basis of “missing evolution” in its genome, detailed measurements of Denisovan and Neandertal admixture into present-day human populations, and the generation of a near-complete catalog of genetic changes that swept to high frequency in modern humans since their divergence from Denisovans.
The Denisova Cave find does not cease to astonish. Located on the other side of the world from what is widely considered to be the centers of human anatomical and behavioral evolution characterized by a wealth of bones and stones (Africa and Europe), Denisova Cave represents, in the human origins discussions, the broad swath of landmass encompassing America, East Asia, Circumpacific zone and the Sahul. Therein human and hominin fossils are rare to non-existent, lithic packages tend to be non-descript and behavioral modernity is not archaeologically visible often until the Holocene. Denisova Cave yielded just three tiny pieces of fossil matter – a phalanx and two molars – but thanks to the ancient DNA that they contained we now have a challenger to many established beliefs about modern human origins.
First, it challenges our deeply-engrained belief that fossils provide a reliable record of human evolution. Denisovans turned out to be a distinct “species” which separated from modern humans by hundreds of thousands of years but with which modern humans apparently interbred. However, up until 2010 (150 years since paleontologists began composing the fossil record) science had no idea they existed. And it would have remained in its state of ignorance if it were not for the newly-developed methods of ancient DNA extraction. Moreover, this new “species” may have ranged from Northeast Asia to Oceania but has remained undetected everywhere but a Siberian cave.
Second, the Denisova Cave find challenges our newly-established belief that, in the genetics of extant human populations, there are nice molecular clusters representing reliable proxies of the real prehistoric divergence of modern human populations as they fanned out from Africa to colonize the rest of the world. More genetically diverse populations, those that contain more of those clusters, are older than less genetically diverse populations, and the seemingly superficial process of visually progressing from counting more clusters in Africa to fewer clusters in Asia, Oceania and America actually represents the way humans colonized the world by taking with them, at every significant junction, a subset of genetic variation of the parent population. However, the Denisova Cave find shows that some modern human populations, namely Papuans, Australians and Island Melanesians, did not just lose ancient African diversity but gained archaic Eurasian diversity through interbreeding. Thanks to Denisovans, it is now clear that taken in separation (ancient) fossils and (modern) are misleading.
Third, the Denisova Cave find (and, of course, ancient Neandertal DNA) opened a conversation regarding whether modern African populations are the products of a unique speciation event within Africa some 200,000 years ago or of admixture between the newly baked non-African humans and African archaics. We have statistical cues that this was likely the case, but we cannot ascertain it unless we find another phalanx or a tooth with recoverable DNA in them. We may find them in a Caucasus cave and the recovered alleles will pop up at high frequencies in South African Bushmen, but we will definitely not find this ancient DNA in Sub-Saharan Africa because DNA does not survive in hot climates. Thanks to Denisovans, it is now clear that the ideal blend between paleobiology and genetics may not be attainable for such a critical continent as Africa. One has to systematically look outside of genetics and paleobiology to reconstruct human prehistory – something that this weblog aims to do.
The new paper by Meyer et al. (2012) takes the Denisova genome to a new level of resolution (31-fold coverage of the ~1.86 autosomal gigabases) and compares it to the genomes of 11 modern human individuals (24- to 33-fold coverage) – a San, Mbuti, Mandenka, Yoruba and Dinka from Africa; a French and Sardinian from Europe; a Han, Dai and Papuan from Asia; and a Karitiana from South America. Predictably, but regrettably, the sample of modern human genomes is heavily biased toward Old World populations, with American Indians comprising 1/5 of the African sample, 1/2 of the European sample and 1/3 of the Asian sample.
One critical outcome of this improved depth of coverage is the finding (p. 3) that Denisovans were very homogeneous genetically (see below, Fig. 5).
“The high quality of the Denisovan genome allowed us to measure its heterozygosity, i.e., the fraction of nucleotide sites that are different between a person’s maternal and paternal genomes. Several methods indicate that the Denisovan heterozygosity is about 0.022%. This is ~20% of the heterozygosity seen in the Africans, ~26-33% of that in the Eurasians, and 36% of that in the Karitiana, a South American population with extremely low heterozygosity. Since we find no evidence for unusually long stretches of homozygosity in the Denisovan genome, this is not due to inbreeding among the immediate ancestors of the Denisovan individual. We thus conclude that genetic diversity of the population to which the Denisovan individual belonged was very low compared to present-day humans.”
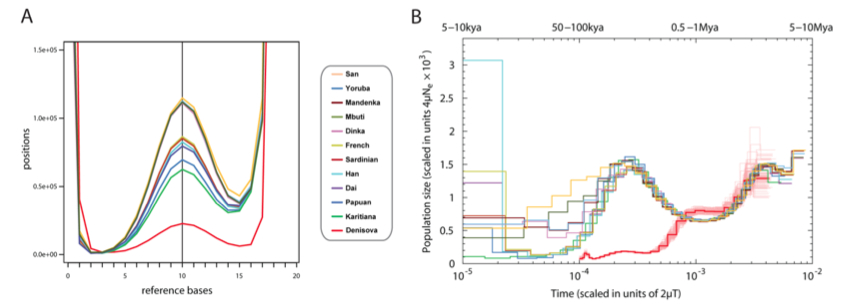
Fig. 5. (A) Heterozygosity shown by the distribution of the number of bases matching the human reference genome at sites sampled to 20-fold coverage. The y-axis is scaled to show the peak representing heterozygous sites in the center. (B) Inference of population size change over time using variation in the time since the most recent common ancestors across the genome shows that Denisovans have had a small population size over the last few hundred thousand years compared with modern humans, but a similar demographic history earlier. The y-axis specifies a number proportional to the population size Ne. The x-axis specifies time in units of divergence per base pair (along the top in years, assuming rates of 0.5×10 -9 to 1.0×10 -9 per year). Thin red lines around the Denisovan curve represent 100 bootstraps, thus showing the uncertainty of the inference.
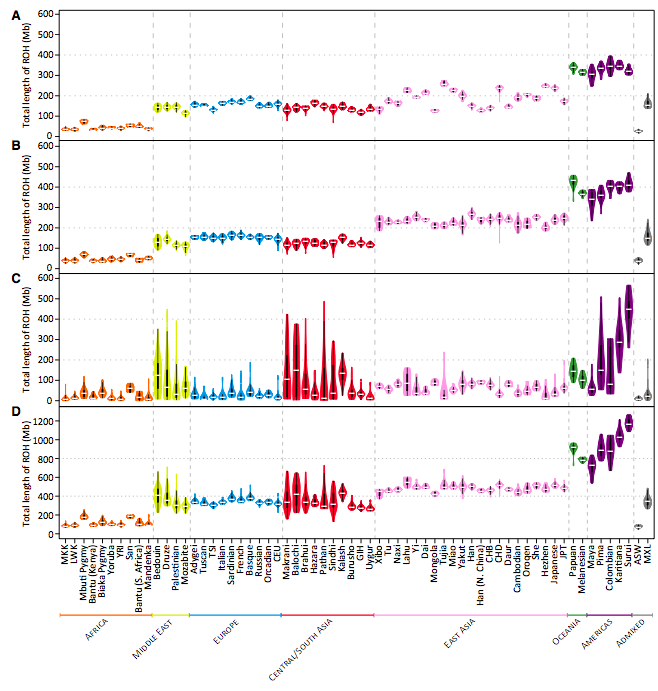
The finding of low heterozygosity among Denisovans undermines one of the central theoretical assumptions of the out-of-Africa thinking, namely that high intragroup allelic diversity indicates great age. The closest modern parallel to the population structure of such a mid-Pleistocene population as Denisovans are not African San and Mbuti (the yellow and green curves on Fig. 5A above) but South American Karitiana (green curve) followed by Papuans (blue curve). There must be other factors (archaic admixture in Africa, mutation rate run-up, etc.) besides age that inflated San and Mbuti intragroup diversity values. Naturally, Karitiana and Papuan demography is more “evolved” than Denisovan, which is consistent with the haploid data showing population expansion across all of the human continental groups. The fact that South American Indians have the lowest heterozygosities among modern human populations has been known before (see below, from Pemberton, Trevor J. et al. 2012. “Genomic Patterns of Homozygosity in Worldwide Human Populations,” American Journal of Human Genetics 91, 280, Fig. 3), but only the high-coverage Denisova sequence provided evidence that this does not need to be taken as an indication of a recent bottleneck that occurred during the peopling of the Americas. Instead, low intragroup diversity and high intergroup diversity are ancient properties of hominin demes best preserved by modern American Indian and Papuan populations. In Africa, only Hadza have retained a “Denisovan” demographic footprint.
There is a perfect alignment between genetic and linguistic data here. America and Papua New Guinea have the highest levels of linguistic diversity in the world. Hadza language is most divergent in the Khoisan family. (Not all linguists accept the Khoisan grouping, but Hadza and San Bushmen share an autosomal genetic component, which indicates that Khoisan languages may be related.) One interesting difference between the Denisovan, on the one hand, and American Indian and Papuan pattern of homozygosity is that in the Americas and in Papua New Guinea runs of homozygosity (ROH) come in all lengths including the longest runs suggesting that in addition to long-term low effective population size there is also cryptic inbreeding affecting the samples and possibly reflecting cross-cousin marriages (see here). Denisova genome, on the other hand, does not show long stretches of homozygosity indicating lack of inbreeding.
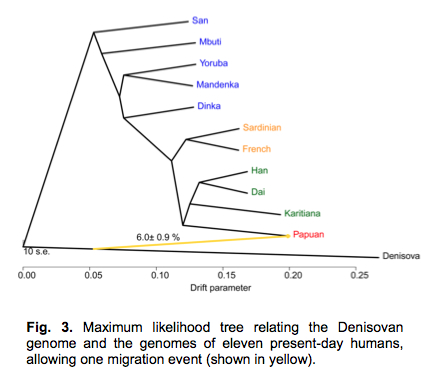
Meyer et al. (2012) applied TreeMix to generate a population divergence tree with admixture. In it, Papuans appear closer to Denisovans followed by Karitiana.
This tree does not factor in Neandertal admixture outside of Africa and African-specific archaic admixture. It is important to take notice of the fact that only the Denisovan-Papuan link represents an actual historical connection. The tree branch connecting the Denisova ancestor (chimpanzee) with San is a hypothetical link generated from the frequencies of chimp-ascertained “ancestral” states in San compared to the other modern human groups. We do not have any African archaic and “anatomically modern” sequences to ascertain the actual phylogenetic continuity between San and chimps to the exclusion of Denisovans, Papuans and Karitiana. Demographic and linguistic data above indicates that the Denisovan-Papuan (and Denisovan-Karitiana) link may be that of common descent, rather than admixture.
A rather enigmatic finding in Meyer et al. (2012, 3) concerns the relationship between Denisovan and Neandertal admixture outside of Africa:
“Interestingly, we find that Denisovans share more alleles with the three populations from eastern Asia and South America (Dai, Han, and Karitiana) than with the two European populations (French and Sardinian) (Z = 5.3). However, this does not appear to be due to Denisovan gene flow into the ancestors of present-day Asians, since the excess archaic material is more closely related to Neandertals than to Denisovans (table S27). We estimate that the proportion of Neandertal ancestry in Europe is 24% lower than in eastern Asia and South America (95% C.I. 12–36%). One possible explanation is that there were at least two independent Neandertal gene flow events into modern humans. An alternative explanation is a single Neandertal gene flow event followed by dilution of the Neandertal proportion in the ancestors of Europeans due to later migration out of Africa. However, this would require about 24% of the present-day European gene pool to be derived from African migrations subsequent to the Neandertal admixture.”
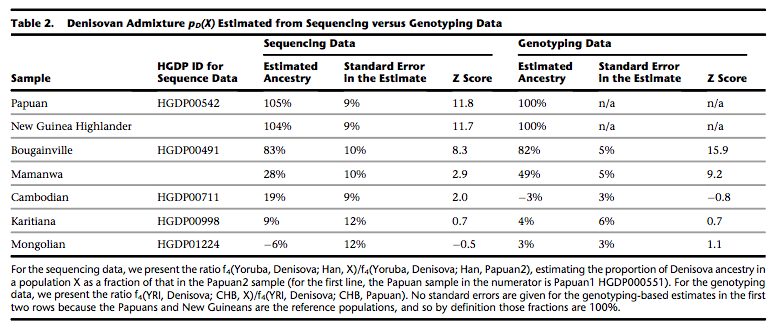
Previous studies detected a small fraction of Denisovan ancestry in Karitiana (same HGDP sequence) compared to a different East Asian population, Mongolians (see below, from Reich et al. 2011. “Denisova Admixture and the First Modern Human Dispersals into Southeast Asia and Oceania,” American Journal of Human Genetics 89, 521; also here).
Apparently, Meyer et al. (2012) do not consider these fractions significant.
Interestingly, the Denisova sequence shows ancestral T allele for EDAR gene (rs3827760) (Meyer et al. 2012, Suppl. Mat., 130). EDAR gene is under strong positive selection in East Asians and American Indians and determines both hair thickness and the incidence of shovel-shaped incisors (see Kimura et al. 2009). Derived C allele is highly frequent in American Indians and East Asians (including Munda-speaking populations in South India, see Chaubey et al. 2011) but is virtually non-existent in Europeans, Africans and Dravidian-speaking South Asians. At the same time shovel-shaped incisors are typical for both Neandertals and Asian Homo erectus. Neandertal shoveling is somewhat different from shoveling found in modern East Asians and American Indians, so it is possible that this highly distinctive dental feature is coded by an altogether different gene in these two species of archaic hominins. But assuming some degree of relationship between Denisovans and Asian Homo erectus, the absence of the derived EDAR allele in Denisovans is puzzling. (For comparison, Denisovans have the mutation at EVC2 with a known association with the Ellis-Van Creveld syndrome in modern humans and may be approaching taurodontism in their dental phenotype (they have enlarged pulp cavity but no fused roots) widely found in Neandertals and, among modern human populations, in Eskimos and American Indians at about 3%.) At the same time, the ancestral allele at EDAR in Denisovans is consistent with modern Papuans’ not having EDAR and its phenotypic manifestations. Considering that Paleoindian skulls tend to cluster with “Australo-Melanesians,” it is likely that Paleoindians had lower frequencies of EDAR gene than their living descendants.
The above-quoted passage from Meyer et al. (2012) suggests that Neandertals and Denisovans being closer relatives than either of them and modern humans, some of the similarity between Denisovans, on the one hand, and Asians and American Indians, on the other, detected in this high-coverage study represents retentions from a common ancestor of Neandertals and Denisovans that made it into modern humans via Neandertals. This, however, creates a rather strained historical scenario. Neandertals that were most densely represented in Western Eurasia first contributed genes equally to all of modern humans, without any bias in favor of Europeans. On a second occasion, they contributed genes exclusively to modern East Eurasians and American Indians. Denisovans, on the other hand, lived in Northeast Asia but contributed genes to modern Papuans to the exclusion of modern Asians and American Indians. The alternative suggested but not favored by Meyer et al. (2012) is that there were two migrations out of Africa – one to Western and Eastern Eurasia, which led to admixture with Neandertals, and the other one to Western Eurasia only after Neandertals had gone extinct. This second African migration diluted Neandertal ancestry in Europeans. But this alternative does not explain why both Neandertal admixture events affected American Indians who supposedly went through a bottleneck that reduced their population size half-way in the direction of the Denisovan bottomline (see above). It also introduces a logic that, if applied consistently, would take us away from talking about the “First Neandertal admixture” in the direction of thinking that the so-called “founding” modern human migration out of Africa to Western and Eastern Eurasia was just gene flow from Africa into an archaic Eurasian population that diluted the concentration of Neandertal and Denisovan genes in all of the present-day human populations outside of Africa.
There is a symmetry between the excess of “Denisovan” alleles in Papuans and the shortage of “Neandertal” alleles in Europeans (both archaic species being closely related to each other), with Northeast Asia, East Asia, Southeast Asia and the New World forming a “hub” with both paleobiological and genetic attestations of an “archaic” hominin source. There is also clear parallelism between the east-to-west decrease in the fraction of autosomal “Neandertal” ancestry and the presence of the “Amerindian” component (mislabeled as “East Eurasian” component by Dienekes) in Western Eurasia. It seems possible that Denisova Cave tells us a story of modern human origins from an East Eurasian hominin, a relative of Neandertals and Denisovans, who speciated into “us” in an isolated refugium such as America and then migrated back into the Old World (see out-of-America II). As early humans were migrating west to Europe and Africa, they lost some of that hominin ancestry and, in Africa, mixed with local archaics who contributed ancestral chimp alleles into a gene pool that had previously been largely composed of derived, or “modern,” as it were, alleles.


A very complete analysis of this really interesting paper. This is the kind of research that brings a more complete understanding of our human origins.
[…] Les scientifiques ont commencé à comparer le génome de la fillette de Dénisova à celui de Néandertal et à ceux de 11 humains modernes, distribués dans le monde ( 26 ). […]