Is Taiwan to Austronesians what America is to Modern Humans?
American Journal of Physical Anthropology 150 (4): 551–564, April 2013
Ascertaining the Role of Taiwan as a Source for the Austronesian Expansion
Sheyla Mirabal, Alicia M. Cadenas, Ralph Garcia-Bertrand, and Rene J. Herrera.
Taiwanese aborigines have been deemed the ancestors of Austronesian speakers which are currently distributed throughout two-thirds of the globe. As such, understanding their genetic distribution and diversity as well as their relationship to mainland Asian groups is important to consolidating the numerous models that have been proposed to explain the dispersal of Austronesian speaking peoples into Oceania. To better understand the role played by the aboriginal Taiwanese in this diaspora, we have analyzed a total of 451 individuals belonging to nine of the tribes currently residing in Taiwan, namely the Ami, Atayal, Bunun, Paiwan, Puyuma, Rukai, Saisiyat, Tsou, and the Yami from Orchid Island off the coast of Taiwan across 15 autosomal short tandem repeat loci. In addition, we have compared the genetic profiles of these tribes to populations from mainland China as well as to collections at key points throughout the Austronesian domain. While our results suggest that Daic populations from Southern China are the likely forefathers of the Taiwanese aborigines, populations within Taiwan show a greater genetic impact on groups at the extremes of the current domain than populations from Indonesia, Mainland, or Southeast Asia lending support to the “Out of Taiwan” hypothesis. We have also observed that specific Taiwanese aboriginal groups (Paiwan, Puyuma, and Saisiyat), and not all tribal populations, have highly influenced genetic distributions of Austronesian populations in the pacific and Madagascar suggesting either an asymmetric migration out of Taiwan or the loss of certain genetic signatures in some of the Taiwanese tribes due to endogamy, isolation, and/or drift.
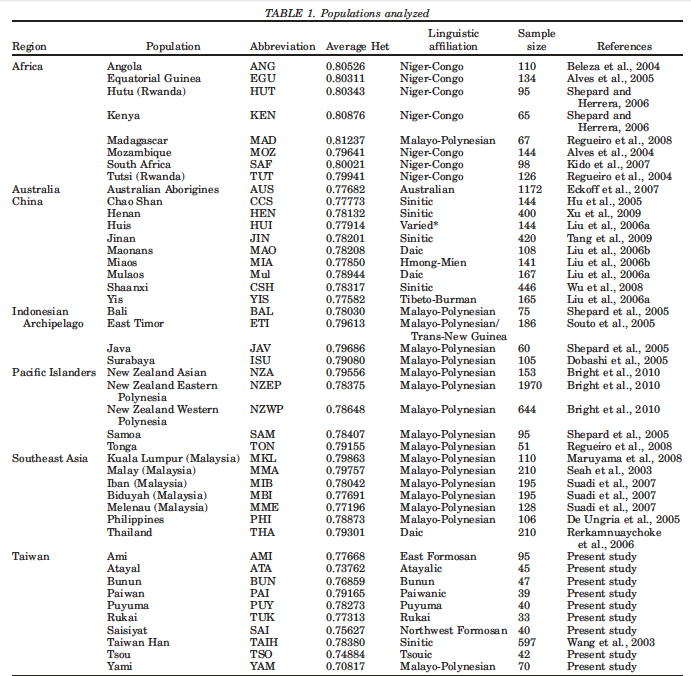
One of the fundamental tenets of the out-of-Africa model has been the argument of genetic diversity: African populations are the most diverse among modern human populations, hence they are the oldest because it takes time to accumulate genetic diversity. The “kitchen logic” behind this argument has had such a powerful impact on both learned academics and not so-learned laymen that nobody has taken trouble to ask: what kind of genetic diversity are we talking about here and whether we have a more direct proof from recent population history that genetic diversity is a function of time. It has been left without a mention that diversity can be measured within populations as well as between populations. This is a textbook analytical and easily quantifiable distinction, which surfaces frequently in “reality of race” debates but has been kept sub rosa in the debates about modern human origins. Mirabal et al. (2013) provide data on intragroup and intergroup genetic diversity for populations involved in one of the better understood human migrations, namely the Austronesian expansion and reaffirm the formative role of Taiwan in it. Their Table 1 (see below) includes average heterozygosities for Taiwanese aborigines next to those of Pacific Islanders and Africans, among others.
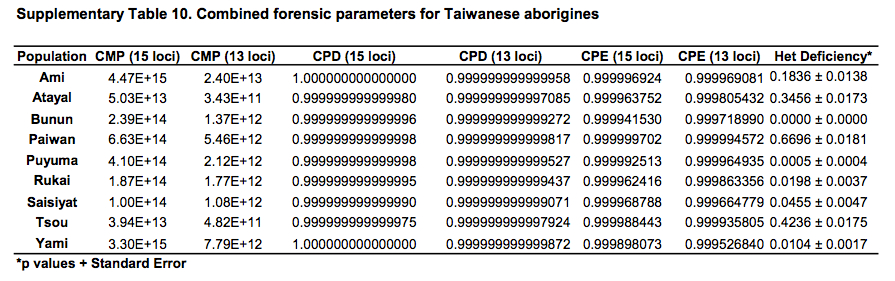
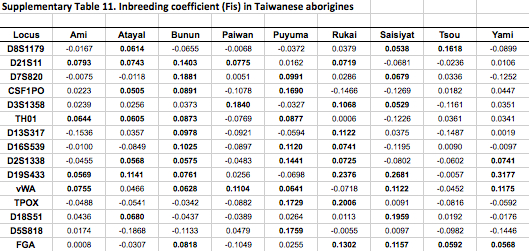
African populations predictably score the highest compared to the rest of the human sample. But it turns out that average heterozygosities among Taiwanese aborigines tend to be lower than those for other Austronesian-speakers in Southeast Asia and Polynesia. Although from the genetic perspective Austronesians originated mostly from Taiwan, higher heterozygosities are found in the derived populations, not in the source ones. Depressed intragroup diversity measures in Taiwanese are also captured by the heterozygosity deficiency and inbreeding coefficient statistics below (Tables 10 and 11).
But where Taiwanese aborigines exceed Southeast Asians and Pacific Islanders is in intergroup diversity as measured by Gst. Mirabal et al. (2013, 559) summarize their findings,
“Genetic distances among Taiwanese aborigines (Gst 50.040195) are such that inter-population variance indices are higher than those found among Pacific populations (Gst 50.026919) which are geographically isolated and contain major biogeographical barriers between them in the form of vast open oceanic distances. Corroborating these statements are the low heterozygosity values (Table 1), severe heterozygote deficiencies (Supporting Information Table 10), high number of loci exhibiting positive inbreeding coefficient correlations (Supporting Information Table 11) and the lowest intrapopulation variance of all the groups of populations analyzed (Hs 50.743899) exhibited by the Taiwanese tribes (Table 2). Altogether, these data suggest that while Taiwanese tribes are genetically different from each other, they are internally highly homogenous, likely due to continued endogamy and genetic drift. These data parallel linguistic divergences between tribes…”
The last point is worth elaborating on further. Taiwanese aborigines show the highest levels of linguistic diversity among Austronesian populations: Formosan languages cluster into as many as 9 first-order branches of the Austronesian family. Formosan linguistic differentiation has been one of the key pieces of evidence for the out-of-Taiwan model of Austronesian origins. Now it becomes clear that the genetic counterpart of this differentiation indicative of a source population is not the intragroup genetic diversity but the intergroup one.
Mirabal et al. (2013, 561) also observed that some Taiwanese populations show greater contributions to Austronesian-speaking populations outside of Taiwan than others. Interestingly, they explain this asymmetry as a function of differential lineage loss within Taiwan:
“We compared each Taiwanese aboriginal population to groups found at the fringes of the Austronesian range in the Pacific and Indian oceans and found
that the Puyuma, Saisiyat and Paiwan are the most genetically influential tribes to modern Austronesian populations and theorize that other Taiwanese tribes may have lost these genetic signatures due to isolation, drift and/or endogamy.”
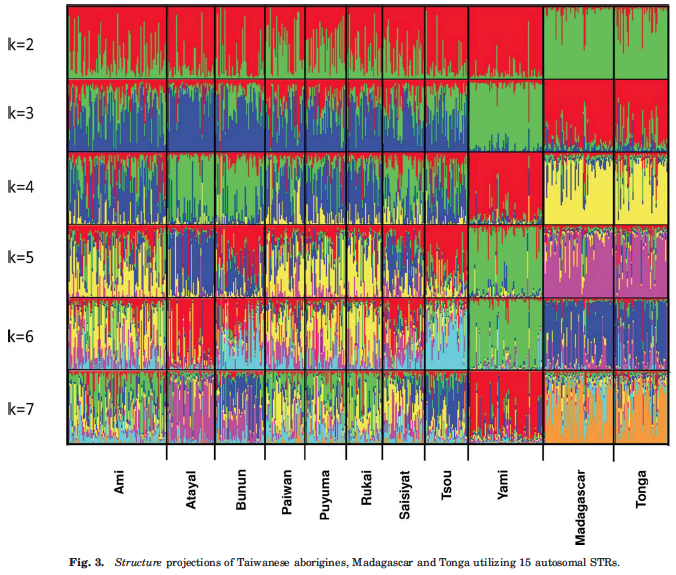
The model of Neolithic population expansion in the Pacific is therefore not the one of serial founder effect linking ancestral and derived populations into a linear chain of intragroup diversity reductions but the model whereby ancestral populations have been losing intragroup diversity to the point that some of them may have lost all of the traces of their founding impact on the colonization of the Pacific. Yami, the population with the lowest heterozygosity (see Table 1 above), also shows the greatest degree of retention (at k=7) of the ancestral (as seen at k=2) Austronesian admixture component (Fig. 3 below, red).
We observe a similar picture on the global scale: Amerindian populations tend to have reduced heterozygosities compared to other modern human populations, especially Africans, but their intergroup diversities are world highest. Tishkoff et al. (2009, 1035) found:
“The proportion of genetic variation among all studied African populations was 1.71% (table S3). In comparison, Native American and Oceanic populations showed the greatest proportion of genetic variation among populations (8.36% and 4.59%, respectively).”
STRUCTURE-kind of analyses tend to depict Amerindians exactly like Mirabal et al.’s (2013) analysis depicted the Yamis – a population that maintains an ancestral component in its purest form (see Fig. 1 one Rosenberg et al. 2002 here). The worldwide distribution of levels of between-population variation corresponds to the phenomenal degree of linguistic differentiation in the New World. This continent harbors 2/3 of world linguistic diversity (excluding Papua New Guinea) as measured by the number of linguistic stocks and families.


Shouldn’t your title read is Taiwan to Austronesians what Africa is to Modern Humans rather than “…What America is to Modern Humans”?
No, the title is exactly what it should be. Africans to non-Africans is nothing like Taiwanese to Polynesians.