A Signal of Neandertal Admixture in Modern Human mtDNA?
Russian Journal of Genetics, September 2013, Vol. 49, No. 9, 975-978
Mitochondrial DNA Polymorphisms Shared Between Modern Humans and Neanderthals: Adaptive Convergence or Evidence for Interspecific Hybridization
Malyarchuk, Boris A.
Abstract
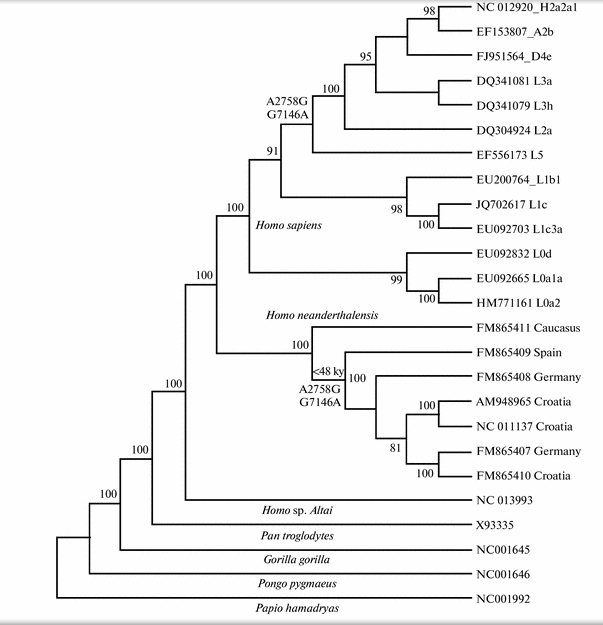
An analysis of the variability of the nucleotide sequences in the mitochondrial genome of modern humans, neanderthals, Denisovans, and other primates has shown that there are shared polymorphisms at positions 2758 and 7146 between modern Homo sapiens (in phylogenetic cluster L2′3′4′5′6) and Homo neanderthalensis (in the group of European neanderthals younger than 48 000 years). It is suggested that the convergence may be due to adaptive changes in the mitochondrial genomes of modern humans and neanderthals or interspecific hybridization associated with mtDNA recombination.
It’s an intriguing publication by a Magadan-based Russian geneticist Boris Malyarchuk (Борис Малярчук) known for his team leadership on several English-language publications devoted to mtDNA variation in Siberia and the New World. Using a novel statistical program, Malyarchuk identified two nucleotide positions identical between European Neandertals and modern humans belonging to the African L2’3’4’5’6 cluster and extra-African M, N and R lineages (under current mtDNA phylogenies M and N are part of L3 and R is part of N) (see below).
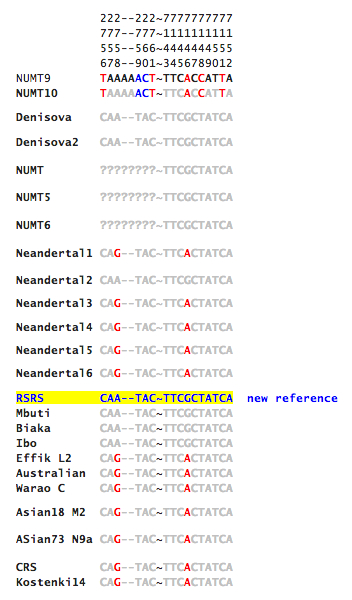
Gisele Horvat has kindly provided me with the actual sequence data around the two sites identified by Malyarchuk as uniquely shared between humans and Neandertals (reproduced below, with permission).
I will continue to update the post as I investigate more around its significance. At face value, if we give preference to the admixture hypothesis over the convergence hypothesis, the data seems to furnish a missing link between a) autosomal data suggesting that “Neandertal admixture” is present in all non-Africans (to a various degree) plus East Africans (see more here); b) and the Y-DNA phylogeny indicating that the main African Y-DNA clade E is a subset of the Eurasian clade CT (xC-F, xD-E). It’s especially notable that the clade showing an introgression from Neandertals encompasses all of African mtDNAs but haplogroups L0 and L1. Hgs L0 and L1 correspond closely to Y-DNA hgs A and B (both sets are highly frequent in Khoisans and Pygmies, are phylogenetically basal and not found outside of Africa).


Neandertals after 48,000 years ago in Western Europe also have greatly restricted mtDNA diversity compared to the earlier sample of Neandertals (Dalén, 2012). Apparently, selective processes increased considerably at the eve of modern human appearance. Also modern human mtDNA seems to have passed through a bottleneck by then. However, did this pressure also result in any noticeable evolution of mtDNA? For this is being implied by convergence. Modern human mtDNA doesn’t seem to have evolved so much since the Tianyuan specimen, and modern human mtDNA is still quite uniform. Neither did modern human mtDNA diverge much further from Neanderthal mtDNA. All the contrary, genetic distances between modern human mtDNA and Neanderthal rather even decreased slightly ever since – especially in the coding region. If this effect was due to some hidden source of Neanderthal mtDNA, this doesn’t explain why Neanderthal remained such a perfect outgroup for the non-coding mtDNA regions of all modern human populations, including even the Tianyuan specimen. I am surprised that scientists already resort to hypothesized explanations that involve cross-species hybridization events for mtDNA while nothing of the sort has ever been observed.
However, excluding hybridization as an explanation there remains another phenomenon, not mentioned by Malyarchuk, that now again glares us in the face. That is, convergence between modern humans and Neanderthal already happened since 100,000 years ago and it happened in the non-coding region! This has been observed comparing the sequenced 123bp string of non-coding HVS-I of the 100 kya Sclandina Neanderthal specimen to more recent Neanderthal. Orlando et al. (2006) dismissed this as random diversity for more ancient Neanderthal (a third hypothesis), without taking in consideration the statistical odds that only Neanderthal mtDNA that was more similar to modern humans survived to the end. Scladina mtDNA was different from more recent Neanderthal in four positions, three of which are shared between modern humans and more recent Neanderthal. I don’t see any clear reason why the older Neanderthal in Belgium should be actually more diverged from the modern human stock than more recent Neanderthal in the same greater region. Actually, I think it more likely that we still have serious dating issues for recent Neanderthal compared to the first modern humans and that we may rather deal with some one time event of evolutionary leapfrogging.
BTW. I am not sure that M. actually intended to refer also to introgression of hybrid Neanderthal mtDNA into modern humans. Probably a hapless effort anyway, in any direction.
Thanks Rokus. Great points! I think Malyarchuk himself is leaning more toward a convergence explanation but he felt obligated to bring the hybridization interpretation up, too, because there’s a lot of evidence of archaic introgression across the human genome and mtDNA/Y-DNA have been somewhat of an exception to the pattern lately.
Do you know if RSRS from Behar et al. 2012 matches the Skladina sequence or the more recent Neandertal ones at those sites identified as polymorphic in Neandertals by Orlando et al.?
What I find interesting, as I tried to point out in the post, is that the 2 matches identified by Malyarchuk are found on the L2’3’4’5’6 clade, but not on the L0 and L1 sequences. The resulting pattern is consistent with Y-DNA and autosomal data. As an extreme interpretation, this would mean that mutations identified by Malyarchuk form a two-site haplotype from which all of non-African and most African lineages are derived.
My general position is that there’s a lot of noise in mtDNA studies and more data and more interpretations are needed to test the current phylogeny. I remember you wrote about the possibility that the current phylogeny is an inverted tree. Do you still hold this opinion?
No, I ran my bit comparisons using rCRS, GenBank number NC_012920. For this little HVR-I string this does not matter, and for the genetic distances I used Blast where this doesn’t matter either.
One of these four mentioned mutations on the Scladina ancestral positions (16258) is still fixed in the mtDNA for modern humans and was polymorphic for the group “more recent Neanderthal”: it showed up in the Neanderthal reference mtDNA genome of Vindija, but it didn’t in Mezmaiskaya. This mutation is the only difference for this 123bp string (positions 16210-16332) between western and eastern Neanderthal. Thus for this single mutation no convergence can be perceived, but instead a partial divergence for the westernmost “more recent Neanderthal”.
Strange enough the shared mutation on position 16243 doesn’t show up in my L0d reference sample (EF184591), though it does in L1a (AF381988) onward. Both other shared mutations on positions 16266, 16291 are fixed in all more recent Neanderthal and all modern humans.
Without counting mutation 16258, the genetic distance of more recent Neanderthal with rCrs is one deletion between 16263-16264 and 10 additional mutations, three of which are polymorph with ancestral values in L0d. Also in the view of leapfrog evolution this confirms an early divergence of L0d from the main branch of modern humans.
Peculiar is that L0d features again the same average genetic distances of non-coding mtDNA, while in the high end of genetic distances for coding mtDNA, even more so than Tianyuan: no more convergence here!
One possible explanation might be that higher genetic distances for the coding region of some ancient populations (Tinayuan, San) are a relict of high effective population sizes and rapid growth that happened once in Africa and Asia. This can’t be observed e.g for Papuan highland mtDNA Q, an offshoot of mtDNA M, I have to take a closer look at this to confirm anything systematic. Maybe here is one reason why I found much of the Denisovan/LM3/Insert “ancestral state” preserved here. However, other such relicts may be found in Africa as well, while some “hidden” markers may be locally missing while present in rCrs. This looks like statistics. So no, I don’t think phylogenetic trees can be simply reversed to explain ancestral details, though apparently some branches evolved at a different pace and thus may give more insight into the true ancestral state of each mutation. I am sure modern human mtDNA is essentially monophyletic, including all African branches, what would mean a single point of divergence in comparison with any outgroup, not so much time before the “more recent Neanderthal” split off – when 16243 was still polymorphic. Taking the available paleogenetic information literally and from a “leapfrog” point of view this could have been somewhere between Scladina and Mezmaiskaya, if so then closer to the latter.
Thanks for this. I’ll read it carefully. In the meantime, I’d appreciate any thoughts on a bigger picture that I put together: http://anthropogenesis.kinshipstudies.org/2013/11/the-end-of-out-of-africa-a-copernican-reassessment-of-the-patterns-of-genetic-variation-in-the-old-world/