Clicks and Genes: Linguistic and Genetic Perspectives on Khoisan Prehistory
Science DOI: 10.1126/science.1227721
Genomic Variation in Seven Khoe-San Groups Reveals Adaptation and Complex African History
Carina M. Schlebusch, Pontus Skoglund, Per Sjödin, Lucie M. Gattepaille, Dena Hernandez, Flora Jay, Sen Li, Michael De Jongh, Andrew Singleton, Michael G. B. Blum, Himla Soodyall, and Mattias Jakobsson
The history of click-speaking Khoe-San, and African populations in general, remains poorly understood. We genotyped ~2.3 million SNPs in 220 southern Africans and found that the Khoe-San diverged from other populations >=100,000 years ago, but structure within the Khoe-San dated back to about 35,000 years ago. Genetic variation in various sub-Saharan populations did not localize the origin of modern humans to a single geographic region within Africa; instead, it indicated a history of admixture and stratification. We found evidence of adaptation targeting muscle function and immune response, potential adaptive introgression of UV-light protection, and selection predating modern human diversification involving skeletal and neurological development. These new findings illustrate the importance of African genomic diversity in understanding human evolutionary history.
While geneticists have been arguing for over 20 years that their data strongly points to an African origin of modern man, the genetic history of modern African populations themselves, prior to the expansion of Bantu-speaking farmers some 5,000 YBP, – remains largely a blank slate. Schlebusch et al. (2012) acknowledge this from the very onset:
“The history of click-speaking Khoe-San, and African populations in general, remains poorly understood.”
They instantly pull out a non-genetic “credentials card” to inform the readers that they focus on click-speaking Khoisan populations, which should immediately signal that the objects of their ~2.3 million SNPs study are outwardly and uncontroversially “archaic.” Otherwise, why would it matter what kind of phonetic peculiarities an object of a genome-wide study possess? No other sounds in the human languages, be it glottal stops or a labiovelars – have the same mouth-watering quality for a geneticist as clicks. (Only one other linguistic moniker, namely “Amerind” stubbornly used by molecular biologists to refer to 140 American Indian language families misclassified by Joseph Greenberg can compete with “clicks” in popularity in geneticists’ circles.) A team of geneticists (Knight et al. 2003. “African Y Chromosome and mtDNA Divergence Provides Insight into the History of Click Languages,” Current Biology 13, 464-473) famously argued that the basal position of Khoisans in haploid phylogenies parallels the phonemic uniqueness of clicks. Together with the fact that clicks are found among supposedly most divergent human populations – Hadza and San Bushmen – this suggested to the researchers that clicks in Khoisan languages must be retentions from the earliest stages in the evolution of the human speech. In 2007, Sarah Tishkoff et al. even put this badge of primitivity into the title of their article “History of Click-Speaking Populations of Africa Inferred from mtDNA and Y Chromosome Genetic Variation.”
Linguists, on the other hand, share nothing of this click hysteria. (And they almost universally reject the Amerind grouping.) Clicks were described for the phoneme inventory of Damin, the secret language of the Australian Lardil tribe (Hale, Kenneth, and David Nash. 1997. “Damin and Lardil Phonotactics,” in Boundary Rider: Essays in Honour of Geoffrey O’Grady, edited by Darrell Tryon & Michael Walsh. Pp. 247-259. Canberra: Australian National University.) indicating that they were clearly recently invented in a strictly localized geographic area and hence they could have been invented relatively recently by Khoisan speakers, too. There are no linguistic reasons to believe that click sounds are primordial in humans (Güldemann, Tom, and Mark Stoneking. 2008. “A Historical Appraisal of Clicks: A Linguistic and Genetic Population Perspective,” Annual Review of Anthropology 37, 93-109).
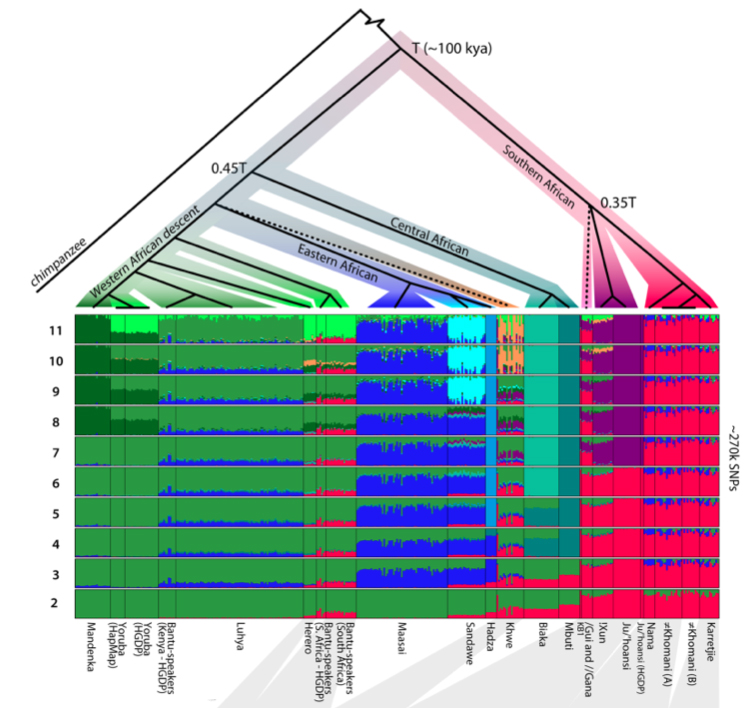
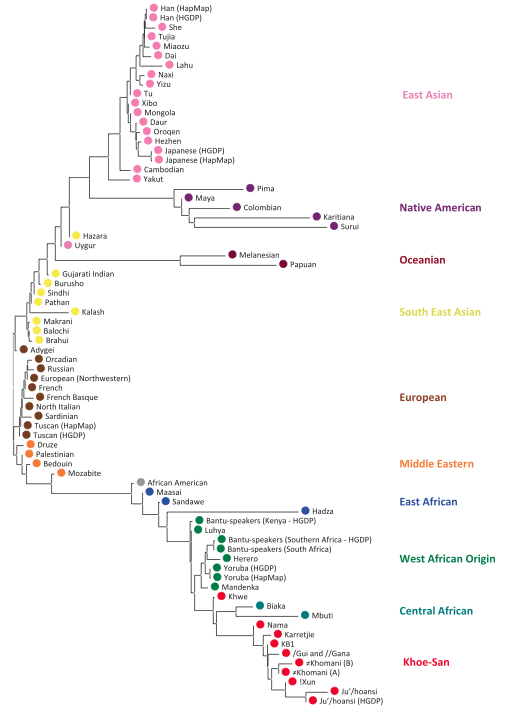
Schlebusch et al. (2012) were boldly marching toward another paper celebrating Khosans’ basal position among modern humans and applied the genealogical concordance model (presumably pressure-tested against possible admixture and drift) to build a usual population tree in which Khoisans diverge earlier than all other humans, including the rest of Sub-Saharan Africans starting with Pygmies (see below). After some 60,000 years of non-descript existence, Khoisans split into Northern and Southern branches at about 25,000-43,000 YBP.
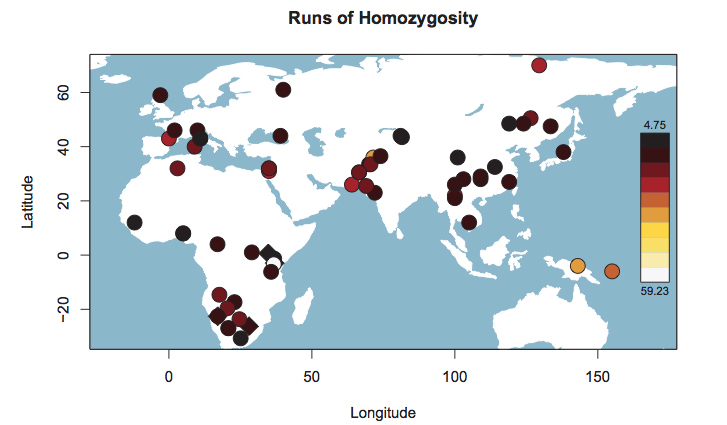
But then they ran into an unexpected result. If the divergence model worked in a clockwork fashion throughout world and African history, one would expect Khoisans to score the lowest values on another measurement, namely Runs of Homozygosity (ROH). ROH are contiguous lengths of homozygous genotypes that are present in an individual due to parents transmitting identical haplotypes to their offspring. A population after a bottleneck contains an excess of ROH, which then progressively break down. Under the hypothesis of a serial founder effect out of Africa with Khoisan populations occupying a basal position in molecular phylogenies, ROH should be the smallest among the oldest populations because these populations had more time to recover from the original bottleneck (see Pemberton et al. 2012. “Genomic Patterns of Homozygosity in Worldwide Human Populations,” American Journal of Human Genetics 91 (2), 275-292). But, surprisingly, it is African farmers (Mandenka) and pastoralists (Maasai) that score the lowest values (see below, table S50 in Suppl. Mat.), with a small, isolated foraging group, Hadza, having the highest ROH values in Africa and such populations as San Bushmen (Ju’hoansi) being in-between.
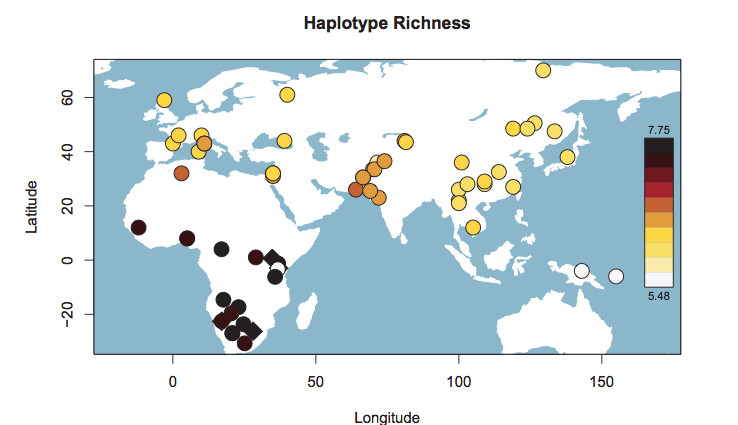
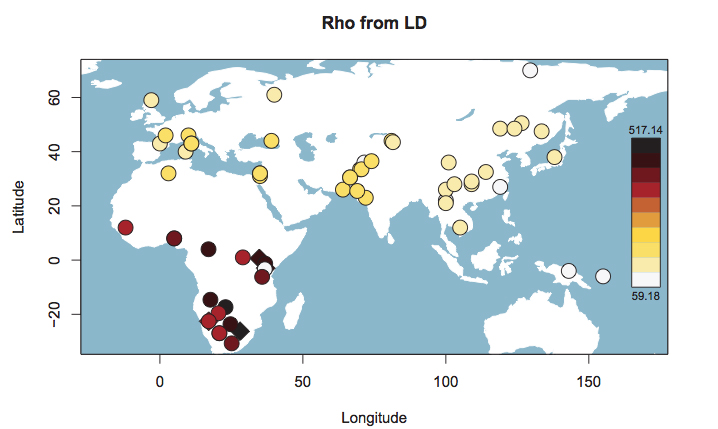
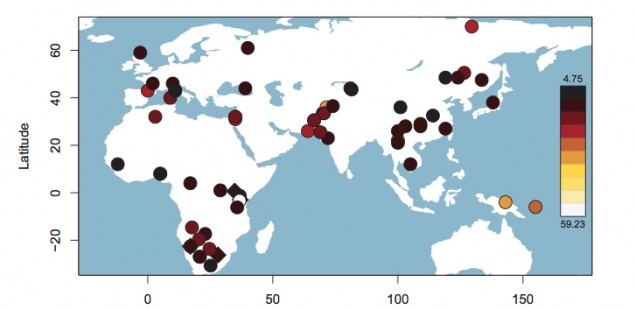
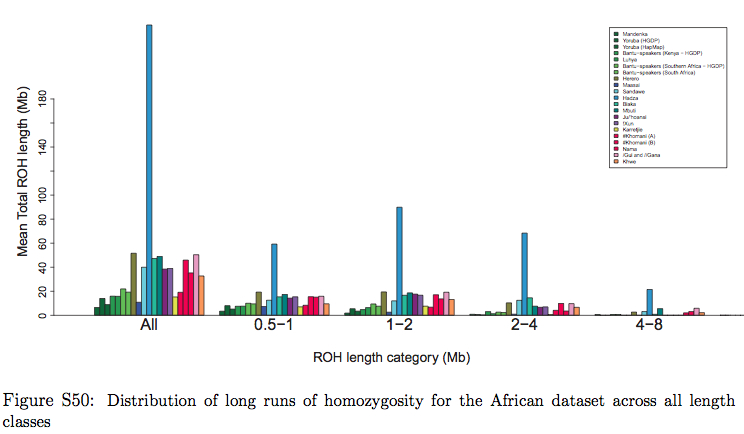 The blue bar in Schlebusch’s Fig. S50 represents Hadza, which fall outside of the typical Sub-Saharan African ROH pattern and are close, on this parameter, to such non-African populations as East Asians, American Indians and Papuans. When looked at from the point of view of haplotype heterozygosity, haplotype richness and linkage disequilibrium (LD), Hadza is a non-African “island” in the middle of East Africa (see below, from Fig. S53 in Schlebusch et al. 2012, Suppl. Mat.).
The blue bar in Schlebusch’s Fig. S50 represents Hadza, which fall outside of the typical Sub-Saharan African ROH pattern and are close, on this parameter, to such non-African populations as East Asians, American Indians and Papuans. When looked at from the point of view of haplotype heterozygosity, haplotype richness and linkage disequilibrium (LD), Hadza is a non-African “island” in the middle of East Africa (see below, from Fig. S53 in Schlebusch et al. 2012, Suppl. Mat.).
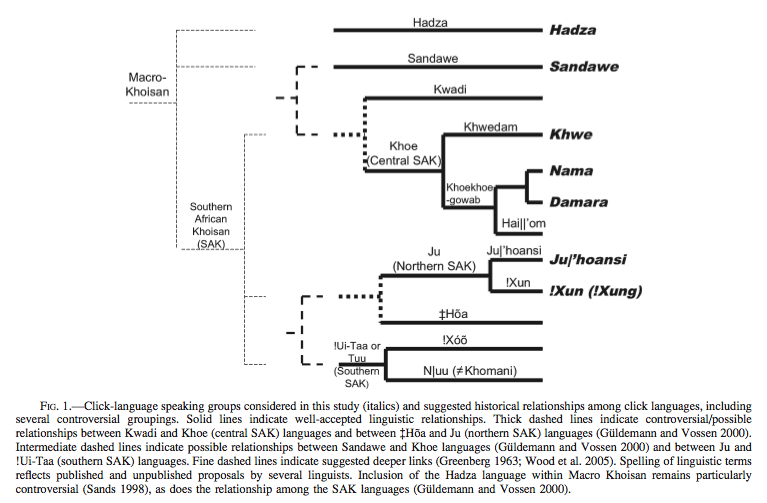
If Hadza is universally recognized as a Khoisan language (some linguists such as Bonnie Sands reject it), which recently received indirect support from autosomal genetics, it would be the earliest offshoot of this family of click-carrying languages. The linguistic relationships – proven and hypothetical – between Khoisan populations are shown below (from Tishkoff et al. 2007).
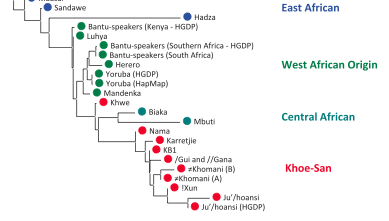
A pullout from Schlebusch et al.’s (2012) Fst-based phylogeny (Fig. S24) shows striking similarities with the phylogeny of Khoisan languages, including Hadza and Sandawe. (Bantu, Yoruba and Mandenka are intrusive here.)
If the linguistic phylogeny is correct and low haplotype heterozygosity, low haplotype richness, high ROH and high LD represent an ancient African genetic condition, Hadza represent a relic of a founding migration of modern humans from Asia to Africa, which eventually led to the formation of South African Khoisans. And not the other way around. (I wonder if “Neandertal admixture” detected in Nilo-Saharan-speaking Maasai in East Africa [e.g., Yotova et al. 2011. “An X-linked Haplotype of Neandertal Origin is Present among All Non-African Populations,” Mol Biol Evol 2011] comes from a substrate population related to Hadza, which Maasai absorbed.) The complete phylogeny from Schlebusch’ et al.’s (2012) Fig. S24 illustrates the whole process.
 American Indian and Papuan populations outside of Africa show the longest branches pointing to two other areas of long-term population isolation in the world comparable and exceeding Sub-Saharan Africa, whereas Adygei, from a highly linguistically diverse West Eurasian refugium, occupy the crest of the (unrooted) phylogeny. Clearly, this unrooted phylogeny tucked away in Supplemental Material to the paper is seriously at odds with Schlebusch et al.’s official, chimpanzee-rooted phylogeny, which misleadingly shows Khoisans splitting off earlier than the rest of humans. Linguists’ staunch hesitation to accept the tempting myth of “clicks as the early human sound” seems to be fully vindicated by a careful examination of genetic data. Realizing what kind of complications their results pose for the overall African origin story, Schlebusch et al. (2012) chose to be diplomatic and elusive:
American Indian and Papuan populations outside of Africa show the longest branches pointing to two other areas of long-term population isolation in the world comparable and exceeding Sub-Saharan Africa, whereas Adygei, from a highly linguistically diverse West Eurasian refugium, occupy the crest of the (unrooted) phylogeny. Clearly, this unrooted phylogeny tucked away in Supplemental Material to the paper is seriously at odds with Schlebusch et al.’s official, chimpanzee-rooted phylogeny, which misleadingly shows Khoisans splitting off earlier than the rest of humans. Linguists’ staunch hesitation to accept the tempting myth of “clicks as the early human sound” seems to be fully vindicated by a careful examination of genetic data. Realizing what kind of complications their results pose for the overall African origin story, Schlebusch et al. (2012) chose to be diplomatic and elusive:
“Thus, these patterns of genetic variation do not localize the origin of modern humans to a single geographic region in Africa, instead they suggest a complex (potentially both recent and ancient) population history within Africa.”
But the concordance between linguistic and genetic pictures seems to be a good indicator of a robust historical inference. For instance, linguistics and genetics walk hand-in-hand in the question of the origins of the South African Nama, or Hottentots. Both disciplines concur that there was a relatively recent (not the founding Hadza-related migration from East Africa) migration of pastoralists from East Africa into the midst of click-speaking populations. In Schlebusch et al. (2012, 2) words,
“The Nama also speak a ‘Central Khoisan’ language and are a Khoe group that traditionally had a pastoralist lifestyle in contrast to the hunter-gatherer lifestyle of the San groups. The Nama showed great genetic similarity to the Southern San groups, such as Khomani and Karretjie (Figs. 1 and 2) and shared a small, but distinct, genetic ancestry component with East African groups, specifically the Maasai, and direct tests showed gene flow from the Maasai to the Nama. This “East African” component was also present at lower levels in the two Khomani groups but basically absent (< 1%) from the !Xun, the Ju/’hoansi and the /Gui and //Gana. The Nama also had a high frequency of a haplotype putatively associated with lactase persistence in the Maasai which was rare in southern African Bantu-speakers, suggesting that lactase persistence in the Nama (50% in adults compared to < 10% in San groups) has an East African origin. These observations support an East African connection for the Nama, and suggest that they originated from a Southern San group that adopted pastoralism with some introgression from an East African group that potentially brought pastoralist practices.”