Intense Admixture: Khoisan Clicks and Khoisan Genes in Southeastern Bantu
European Journal of Human Genetics 2012, 1-7 DOI:10.1038/ejhg.2012.192
Genetic Perspectives on the Origin of Clicks in Bantu Languages from Southwestern Zambia
Chiara Barbieri, Anne Butthof, Koen Bostoen, and Brigitte Pakendorf
Some Bantu languages spoken in southwestern Zambia and neighboring regions of Botswana, Namibia, and Angola are characterized by the presence of click consonants, whereas their closest linguistic relatives lack such clicks. As clicks are a typical feature not of the Bantu language family, but of Khoisan languages, it is highly probable that the Bantu languages in question borrowed the clicks from Khoisan languages. In this paper, we combine complete mitochondrial genome sequences from a representative sample of populations from the Western Province of Zambia speaking Bantu languages with and without clicks, with fine-scaled analyses of Y-chromosomal single nucleotide polymorphisms and short tandem repeats to investigate the prehistoric contact that led to this borrowing of click consonants. Our results reveal complex population-specific histories, with female-biased admixture from Khoisan-speaking groups associated with the incorporation of click sounds in one Bantu-speaking population, while concomitant levels of potential Khoisan admixture did not result in sound change in another. Furthermore, the lack of sequence sharing between the Bantu-speaking groups from southwestern Zambia investigated here and extant Khoisan populations provides an indication that there must have been genetic substructure in the Khoisan-speaking indigenous groups of southern Africa that did not survive until the present or has been substantially reduced.
This is another recent paper that discusses the genetics and linguistics in South Africa. Barbieri et al. (2012) find a good fit between genetic and linguistic data when it comes to such aspect of the history of click languages in Africa as their borrowing by several Bantu languages. If linguists have always been on the fence about adopting the idea that clicks are living phonetic fossils, they readily accept the fact that clicks can be borrowed. The borrowing of clicks by non-Khoisan languages is very well attested in Africa, and indirectly this serves as another reminder to geneticists that clicks are normal human sounds subject to the same evolutionary logic of multiple emergence, decay and diffusion.
It is well known that among Bantu-speakers Zulu and Xhosa have clicks. Barbieri et al. (2012) describe another, lesser known cluster of Bantu languages that apparently borrowed click sounds from Khoisan languages.
“These are spoken in a small contiguous area encompassing southeastern Angola, southwestern Zambia, northwestern Botswana, and northeastern Namibia, and belong to different subgroups of the Bantu family. In the Botatwe subgroup, clicks are found only in Fwe, being absent from the closely related languages Shanjo, Totela, and Subiya and the more distantly related Tonga; in the Luyana subgroup, clicks are found in Mbukushu, but are absent from its close relative Kwamashi” (Barbieri et al. 2012, 1).
Khoisan populations are genetically very specific, hence it is easy to detect Khoisan admixture among Bantu-speakers. Khoisan admixture was detected in all Botatwe and Luyana speaking groups (~29% of Khoisan-specific mtDNA haplogroups L0d and L0k7 and ~5% of Y-chromosomal haplogroup A-M51) regardless of whether these groups borrowed clicks or not. Barbieri et al. (2012, 1) identify several demographic models under which admixture between Khoisans and southeastern Bantu could occur:
“Apart from their independent innovation in the Bantu languages, which is highly unlikely, there are three probable pathways by which clicks might have entered the Southwest Bantu languages that have them: (1) through superficial ‘culture contact’ in which Bantu speakers borrowed words containing clicks from Khoisan languages without further intimate contact; (2) through language shift, in which entire groups of Khoisan speakers, both men and women, gave up their original language in favor of a Bantu language, transferring some words and sounds to the new language in the process; or (3) through intermarriage between Bantu speakers and Khoisan speakers. If the sociocultural situation in prehistoric times was similar to that of the present, this intermarriage is likely to have been sex-biased, with Khoisan- speaking women marrying Bantu-speaking men, but not the opposite.”
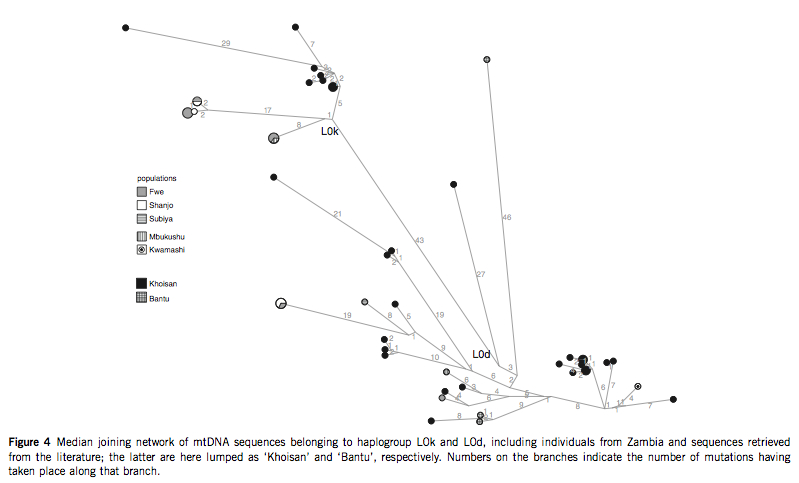
They favor model 3 and propose that the borrowing of clicks from Khoisans (Ju or Khwe) into southeastern Bantu followed sex-biased admixture on the maternal line. (Interestingly, if linguistic divergence was shown to be strongly correlated with Y-DNA patterns, linguistic borrowing can apparently be associated with admixture in mtDNA.) This admixture must have been intense, the authors note, because of the amount of divergence between the Khoisan-derived sequences in Fwe: two of the L0k sequences are separated by 27 mutations and two L0d sequences are separated by 56 mutations.
“To accumulate this amount of divergence from a single shared ancestor per haplogroup would take more than a thousand generations, whereas Bantu speakers arrived in Zambia only around 40 generations ago” (Barbieri et al. 2012, 6).
The intense admixture scenario for the emergence of clicks in southeastern Bantu languages makes sense considering the known difficulties with which adults learn clicks. It must have taken a large number of click-speaking women to enter Fwe to introduce a set of whole new phonemes with complex articulation. But this straightforward scenario faces an unexpected problem: there is a) a lack of haplotype sharing between the Fwe and Khoisan populations, b) the branches, which lead to the Zambian Bantu L0k and L0d haplotypes, are very long (see Figure 4 below), and c) the pattern of frequencies of the two lineages is reversed from the pattern observed in Khoisan and other Khoisan-admixed populations. (In Zambian Bantu, L0k is more frequent than L0d.)
Barbieri et al. (2012) approach this conundrum from several different perspectives: a) data coverage may still be poor for Khoisans and the intermediary lineages may be detected in the future through better sampling; b) drift may have erased these lineages from the Khoisan; c) a Khoisan-like population that was the donor of these divergent lineages was completely replaced by the migrating Bantu; or, d) the ancestral Khoisan population was subdivided.
Intense genetic admixture does not, by itself, predict that the Bantu population, which has a substantial amount of Khoisan genes, will have clicks. The authors bring up the case of Shanjo who show the same level of Khoisan-derived L0k and L0d haplotypes as Fwe but who lack clicks. It is possible that the L0k and L0d haplotypes entered Shanjo via Fwe and not directly from the Khoisan because L0k and L0d haplotypes are similar in Fwe and Shanjo. But Shanjo and Fwe don’t seem to share Bantu haplotypes suggesting that these haplotypes came directly from the Khoisan and that the presence or absence of clicks in non-Khoisan language is determined by sociocultural reasons.
Barbieri et al. (2012) conclude:
“The precise modality of the contact between the ancestors of the Fwe and Khoisan-speaking populations is hard to elucidate, but ultimate replacement of the Khoisan group by the Bantu-speaking community coupled with some female-biased admixture is the most plausible scenario.”
This does seem to be a very reasonable and predictable scenario. Alternatively, exactly the same scenario is postulated by scholars on the basis of genetic data in the case of African Pygmies and Bantu populations. But the linguistic outcome of these two kinds of interactions – between foraging Khoisan and farming Bantu and between foraging Pygmies and farming Bantu is radically different. At least, according to the most widely spread belief (which I challenge for various reasons), as Bantu populations fanned out from their homeland in West Africa and encountered short-statured foraging populations in the African tropical forest, there were some 20 independent cases of language shift among Pygmy populations leading to their (arguably) complete loss of their original tongues and adoption of various Bantu dialects. Although there is some debate (mostly driven by Serge Bahuchet) whether different Bantu-speaking Pygmy groups share a substrate vocabulary related to hunting and gathering, there is absolutely no evidence that the languages of Bantu farmers were in any way impacted by the phonetics, lexicon or grammar of the Pygmy groups they presumably replaced. Similarly, there are no Khoisan-looking (e.g., having lighter skin, sporting an epicanthus, etc.) populations speaking Bantu languages.
The genetic parallels between Pygmy-Bantu and Khoisan-Bantu “admixture” are striking. Just like Khoisans and Fwe occupy different branches of the same deeply rooting hg L0, Western Pygmies and Western Bantu share various lineages within hg L1c. In the case of Pygmies, some mtDNA researchers suggest both “deep common ancestry” between Pygmies and Bantu going back to 70,000 YBP and asymmetric, sex-biased gene flow that took place much later.
“The occurrence of various L1c clades in both AGR [Bantu agriculturalists. – G.D.] and PHG (Pygmy hunter-gatherers. – G.D.], the particularly high frequencies of this Hg among all western HG and the early coalescence age of the autochthonous L1c1a in CA (57,100 YBP) suggest that the maternal gene pool of the ancestors of contemporary AGR and PHG was dominated by the various L1c clades (probably including Hgs now extinct). Two populations arose from this presumed ancestral population: the modern AGR population, which includes various L1c clades (L1c1a, L1c1b, L1c1c, L1c2–6, etc.), and the western PHG population, in which L1c1a is the only surviving clade. The Pygmies must have split from this ancestral population no more than 73,800 years ago, when L1c1a began to diverge from L1c1 (Fig. 1). A long period of isolation (i.e., genetic and/or cultural) must then have occurred, accounting for the phenotypic differences characterizing PHG groups.” (Quintana-Murcdi et al. 2008. “Maternal Traces of Deep Common Ancestry and Asymmetric Gene Flow between Pygmy Hunter-Gatherers and Bantu-Speaking Farmers,” PNAS 105 (5), 1599-1600)
In the case of Khoisans and Fwe, Barbieri et al. (2012) are not considering “deep common ancestry” to explain the peculiar sharing and divergence of the deeply rooting L0k and L0d haplotypes. Precisely because linguistic evidence unambiguously points to an “admixture” scenario on the genetic side. In the case of Pygmies, the fact that, linguistically speaking, Pygmies are Bantu allows one to see behind haplotype sharing between Pygmy and Bantu populations a signal of common descent, not just admixture.