Molecular Variance Across Genetic Systems in Modern Humans and Their Kinship Structures
Gisele Horvat has kindly pointed me to this new pre-print. This post is cross-posted at www.kinshipstudies.org.
Global patterns of sex-biased migrations in humans
Chuan-Chao Wang, Li Jin, Hui Li.
Abstract
A series of studies have revealed the among-population components of genetic variation are higher for the paternal Y chromosome than for the maternal mitochondrial DNA (mtDNA), which indicates sex-biased migrations in human populations. However, this phenomenon might be also an ascertainment bias due to nonrandom sampling of SNPs. To eliminate the possible bias, we used the whole Y chromosome and mtDNA sequence data of 491 individuals from the 1000 Genomes Project Phase I to address the sex-biased migration dispute. We found that genetic differentiation between populations was higher for Y chromosome than for the mtDNA at global scales. The migration rate of female might be three times higher than that of male, assuming the effective population size is the same for male and female.
The paper contains a rundown of inter- and intragroup diversity values for both mtDNA and Y-DNA on a worldwide scale (see below).
As far as I know, this is the only paper that has pooled it all together for the two haploid systems. This is consistent with the classic observation that most of the variation found among modern humans is concentrated within human groups. But it’s noteworthy that the continents differ in the degree in which this pattern is manifested. Some continents and populations buck the general human trend by accumulating more variation between groups than others. The notable pattern in this data, as Wang et al. (2013) point out, is that
“the between-population component of genetic variation was slightly higher for mtDNA than for the Y chromosome in America and Africa (by 5~6 times), but not in Europe and Asia. In Europe and Asia, between-population component of genetic variation was about 10~20 times higher for Y chromosome than for the mtDNA.”
What this pattern likely means is that Amerindian and Sub-Saharan African populations have been more consistently matrilocal/uxorilocal/matrilineal, while populations outside of Africa and America more consistently patrilocal/virilocal/patrilineal. Wang et al. (2013) confirm this interpretation and refute the conclusion reached by Wilder et al. 2004 (“Global Patterns of Human Mitochondrial DNA and Y-Chromosome Structure Are Not Influenced by Higher Migration Rates of Females versus Males,” Nat Genet 3610, 1122-1125) that there is no correlation between social structure and sex-biased migration rates, on the one hand, and molecular variation at haploid loci, on the other.
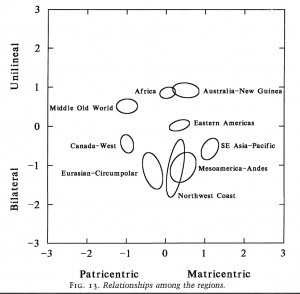
Wang et al.’s data and interpretations are consistent with the worldwide data from social anthropology on the relative frequencies of matricentric vs. patricentric social structures. Burton et al. (“Regions Based on Social Structure,” Current Anthropology 37 (4), 1996, 93) define matri- and patricentric forms of social organization in the following way:
“Matricentric social organization traits include localized or dispersed matrilineal groups, matrilocal or uxorilocal residence, monogamy and the absence of marriage exchange. Matricentric societies tend to organize kinship groups around women through matrilocal or uxorilocal residence or through matrilineal kinship groups. Patricentric social organization traits include nomadic or seminomadic settlement patterns, clan communities, localized or dispersed patrilineal groups, patrilocal residence, polygyny and bridewealth payments. Hence, patricentric societies tend to organize kin groups around men, through patrilocal residence, patrilineal descent or polygyny.”
The worldwide distribution of matri- vs. patricenrtic structures is skewed between Africa, the Pacific and America, on the one hand, and Eurasia, on the other (see below, from Burton et al. 1996, 109), which is consistent with Wang et al.’s data from mtDNA and Y-DNA. North American Indian societies is the only exception from this pattern – they fall on the patricentric side.
My own kin-terminological data shows the paucity of “Crow” terminologies (highly correlated with matrilineal descent) in Western, Eastern Eurasia, Southeast Asia and Australia and their relative salience in Sub-Saharan Africa and America (as well as in the Pacific). “Omaha” systems (highly correlated with patrilineal descent) are the only descent-skewed terminologies found in those areas.
Now that we have global molecular variance data for the haploid loci, we can compare them with the same statistics derived from autosomal loci (see below, from Tishkoff et al. 2009. “The Genetic Structure and History of Africans and African Americans,” Suppl.Mat., Table S3).
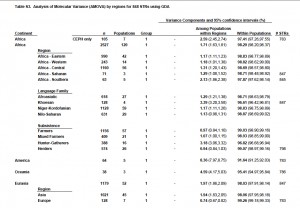 Tishkoff et al.’s data is broken down by continent, by population and by economic system. Notably, African hunter-gatherers in general and Khoisans in particular show a different pattern from African pastoralists and farmers: they are closer to Amerindian and Oceanic populations in having a greater fraction of their variance come from inter-group values. This is consistent with the more specific data that we have on Hadza, linguistically the most divergent among Khoisan languages, that shows that Hadza has less intragroup variation than other Sub-Saharan Africans and is consequently closer to the Amerindian pattern of variance (see here and here). In turn, the Amerindian pattern of variance (more intergroup diversity, less intragroup diversity) is similar to the one found in Denisovans.
Tishkoff et al.’s data is broken down by continent, by population and by economic system. Notably, African hunter-gatherers in general and Khoisans in particular show a different pattern from African pastoralists and farmers: they are closer to Amerindian and Oceanic populations in having a greater fraction of their variance come from inter-group values. This is consistent with the more specific data that we have on Hadza, linguistically the most divergent among Khoisan languages, that shows that Hadza has less intragroup variation than other Sub-Saharan Africans and is consequently closer to the Amerindian pattern of variance (see here and here). In turn, the Amerindian pattern of variance (more intergroup diversity, less intragroup diversity) is similar to the one found in Denisovans.
If molecular variance at the haploid loci points to the impact of residence and unilineal descent on the genetic structure of human populations, the sociodemographic reality behind the autosomal variance statistics is that older human populations associated with a more ancient subsistence pattern were smaller, more isolated from each other and affected by inbreeding and genetic drift to a larger degree than more recent populations associated with more recent subsistence patterns.