Ancient Ust’-Ishim DNA as Seen From the Americas
Nature 514, 445–449 (23 October 2014) doi:10.1038/nature13810
Genome sequence of a 45,000-year-old modern human from western Siberia
Qiaomei Fu, Heng Li, Priya Moorjani, Flora Jay, Sergey M. Slepchenko, Aleksei A. Bondarev, Philip L. F. Johnson, Ayinuer Aximu-Petri, Kay Prufer, Cesare de Filippo, Matthias Meyer, Nicolas Zwyns, Domingo C. Salazar-Garcıa, Yaroslav V. Kuzmin, Susan G. Keates, Pavel A. Kosintsev, Dmitry I. Razhev, Michael P. Richards, Nikolai V. Peristov, Michael Lachmann, Katerina Douka, Thomas F. G. Higham, Montgomery Slatkin, Jean-Jacques Hublin, David Reich, Janet Kelso, T. Bence Viola, and Svante Paabo.
We present the high-quality genome sequence of a ~45,000-year-old modern human male from Siberia. This individual derives from a population that lived before—or simultaneously with—the separation of the populations in western and eastern Eurasia and carries a similar amount of Neanderthal ancestry as present-day Eurasians. However, the genomic segments of Neanderthal ancestry are substantially longer than those observed in present-day individuals, indicating that Neanderthal gene flow into the ancestors of this individual occurred 7,000–13,000 years before he lived. We estimate an autosomal mutation rate of 0.4 × 10−9 to 0.6 × 10−9 per site per year, a Y chromosomal mutation rate of 0.7 × 10−9 to 0.9 × 10−9 per site per year based on the additional substitutions that have occurred in present-day non-Africans compared to this genome, and a mitochondrial mutation rate of 1.8 × 10−8 to 3.2 × 10−8 per site per year based on the age of the bone.
Link Ancient.Asia.Fu-2014(Ust-Ishim)
The long-awaited ancient DNA study of the 45,000-year-old modern human individual from north of Ust’-Ishim in Siberia is finally out. The paleogenomics-induced revolution in our understanding of modern human origins continues on. Here’s a summary of Fu et al.’s principal and uncontroversial findings:
1. UI belongs to basal mtDNA R and Y-DNA K haplogroups.
2. UI is closer to modern and ancient non-Africans than to modern Africans.
3. Among modern non-Africans, UI is closer to East Eurasians than to ancient and modern West Eurasians.
4. Among West Eurasians UI is closer to ancient West Eurasians (Mal’ta at 24,000 YBP and La Brana at 7,000 YBP) than to modern West Eurasians.
5. UI has longer stretches of Neandertal DNA than other non-Africans suggesting that, as befits an ancient sample, it’s closer to the putative “admixture” event between non-Africans and Neandertals than modern non-Africans and it had less time to break down the introgressed chunks of archaic DNA.
6. UI is closer to Neandertals than modern East Eurasians and East Eurasians (consistently with earlier findings) are closer to Neandertals than West Eurasians.
7. UI’s level of heterozygosity falls within the range of modern West Eurasian variation. UI is not as heterozygous as modern Africans but it’s significantly more heterozygous than Eurasian hominins (Altai Neandertals and Denisovans) and modern Amerindians. This suggests that UI was an admixed population and consequently it’s equally close to Onge (“Eastern non-Africans”), MA-1 (“Ancient North Eurasians”) as well as La Brana (“West Eurasians”).
8. Just like the Mal’ta (MA-1) sample from 24,000 YBP, UI shows a diverse set of population components. A minor one (BLACK) is found in modern Amerindians (same as MA-1), while BLUE and MAGENTA are shared with Sub-Saharan Africans (not present in MA-1). Unlike MA-1, UI shows the East Asian component (YELLOW) (see below; the MA-1 data is from Raghavan et al. 2013, SI, 55).
9. Just like MA-1, UI shows an ancestral A allele of the EDAR gene. The derived EDAR G allele is a molecular source of a diverse set of East Asian phenotypical characteristics, including hair texture and dental shoveling. Other genetic signatures of the “Mongoloid” phenotype (e.g., oculocutaneous albinism II on chromosome 15 [OCA2] gene coding for light skin in East Asians) are also missing on UI. This supports the hypothesis of a late, intrusive nature of EDAR-A in East Asia and its origin in the Americas.
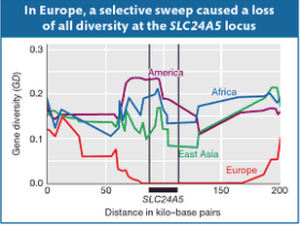
10. Similarly, one of the most common ancestry-defining markers, namely solute carrier family 24 member 5 (SLC24A5) gene that, among Europeans, is almost fixed in the derived G allele in the rs1426654 SNP and that codes for European light skin is not present in UI. Sometime in the Holocene, a famous selective sweep eliminated all of the diversity at SLC24A5 in Europeans. Notably, Amerindians show the highest gene diversity at this locus followed by Africans, East Asians and, finally, Europeans (see below, Fig.18-23 in Solutions Manual for Introduction to Genetic Analysis. London: Macmillan, 2010, 670). This suggests that Europeans inherited the tail end of a much more ancient selective process that affected all of the Old World populations.
From the out-of-America II perspective, modern Amerindians represent the best surviving example of a founding modern human population that speciated from an East Eurasian hominin species and recolonized the Old World between 100,000 and 50,000 YBP. Considering that UI is the earliest modern human DNA presently available and, importantly, the one that’s closer to Neandertals than modern humans, it holds special significance for the testing of out-of-America II.
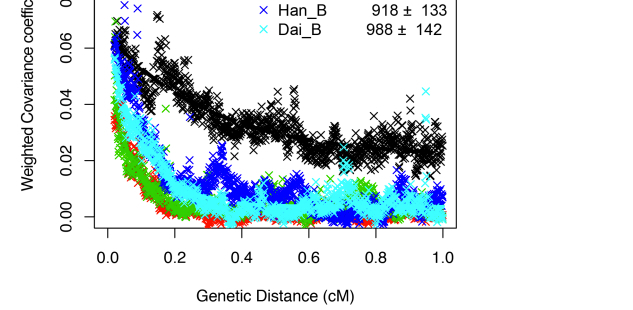
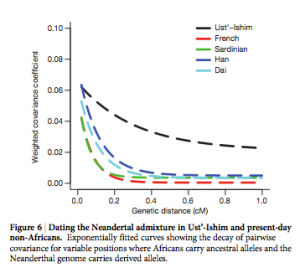
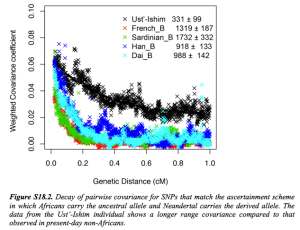
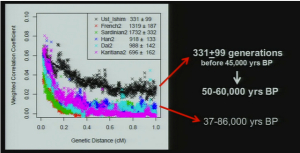
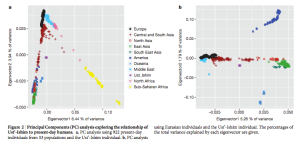
Clearly, Amerindians throw a wrench into the mainstream interpretation of modern human molecular evolution. Fu et al. (2014) went as far as erasing Amerindians from some of their key data plots. The two graphs below – one is a more thorough version from Supplemental Information and the other is a sketch that appears in the main article – demonstrate the greater proximity of UI to Neandertals, and one can observe the gradual progression from UI to East Eurasians and then to West Eurasians. As John Hawks writes,
“Shared alleles with Neandertals are co-inherited across longer distances in the Ust’-Ishim genome than in living people. That is, there is greater linkage disequilibrium between Neandertal-derived alleles in the Ust’-Ishim genome. That indicates that Ust’-Ishim lived much sooner after its Neandertal ancestors than we do.”
Notably, Amerindians are not shown on these graphs. However, an earlier version of the same data plot presented by one of the co-authors, Svante Paabo, at a conference (see the reel at approximately 1:01:03) earlier this year does contain Amerindians.
Karitiana follow UI in the number of generations that have supposedly elapsed since the putative admixture event with Neandertals. But Karitiana is 45,000 years younger than UI! What this data plot shows is that Karitiana closer to Neandertals than Han or Dai and that the decay of Neandertal chunks must have occurred at a slower mutation rate in the Americas than elsewhere. No surprise, Fu et al. (2014) decided to sweep this “inconvenient truth” under the carpet and not show Karitiana at all.
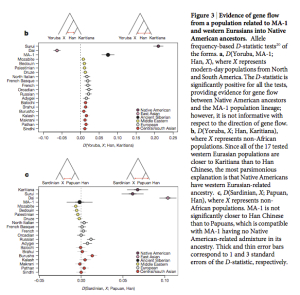
Interestingly, Fu et al. (2014, 448-9) found that UI “is not more closely related to the Onge from the Andaman Islands (putative descendants of an early coastal migration) than he is to present-day East Asians or Native Americans (putative descendants
of a northern migration).” This comes from the D statistic runs that assess the proportion of shared derived alleles between 3 human populations using chimpanzee as an outgroup (see below). 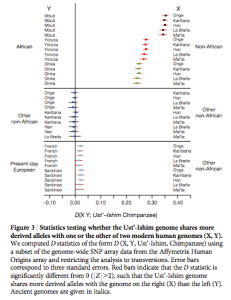 The D statistic even shows a slight gravitation of UI to Onge and Han compared to Karitiana (Fig. 3 on the left, upper band). Fu et al. (2014) don’t deem this significant. But what is significant is the combination of two data points: Onge and Han are closer to UI than Karitiana but Karitiana is closer to West Eurasians than Onge or Han.
The D statistic even shows a slight gravitation of UI to Onge and Han compared to Karitiana (Fig. 3 on the left, upper band). Fu et al. (2014) don’t deem this significant. But what is significant is the combination of two data points: Onge and Han are closer to UI than Karitiana but Karitiana is closer to West Eurasians than Onge or Han.
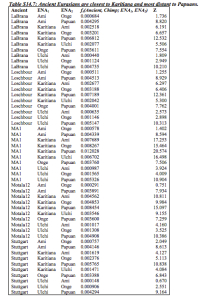
The latter data pattern is supported by a number of recent studies (from Patterson et al. 2012 on) that show that Amerindians are closer to West Eurasians (both ancient, such as Mal’ta and La Brana, and, in the light of Ust’-Ishim’s East Eurasian skew, importantly, modern) than East Eurasians. Below are two data tables – one from Ragahvan et al. 2013 and the other one from Lazaridis et al. 2014 showing a consistent trend toward the greater proximity of European populations to Amerindians compared to East Eurasians.
So, the possible reason why the D statistic does not show the greater proximity between UI and Amerindians compared to East Eurasians (clearly seen in the proportion of Neandertal-related chunks of the genome (see above)) is likely the special affinity between Amerindians and West Eurasians and hence the membership of Amerindians in a phylogenetic node that predated the split between West Eurasians and East Eurasians.
As we have seen above, among Fu et al.’s (2014) principal and uncontroversial findings is the observation that UI is closer to East Eurasians (including Amerindians and Papuans) than to West Eurasians. So, while there’s no evidence for the putative earlier coastal migrants out of Africa, there’s evidence for the split between West Eurasians and East Eurasians. Notably, this pattern cannot be explained as product of a later gene flow from Sub-Saharan Africa.
“This is unlikely to be an artifact of the low levels of gene flow between Yoruba and West Eurasians that have previously been documented, as when we recomputed the statistic with human-chimpanzee common ancestor (CA) or only chimpanzee as the
outgroup instead of the Yoruba, we obtain similar results (Table S11.2).” (Fu et al. 2014, SI, 67).
The authors explain their paradoxical finding as the presence, in West Eurasians, of a putative ancient “Basal Eurasian” ancestry component (Lazaridis 2014) from the time before the separation of UI and East Eurasians. UI does not have it (if it did, UI would have been equidistant from West and East Eurasians), while Europeans received it as a result of a Neolithic migration of farmers from the Middle East. In the Pleistocene, Europeans were not genetically different from East Eurasians (they were all part of the so-called “Ancient North Eurasian” (ANE) population), but then Neolithic farmers brought with them a very ancient population component not found in modern Sub-Saharan Africans but existing at appreciable frequencies in modern Middle Eastern and North African populations.
This said, Fu et al. (2014, 448) assume – in reiteration of a pre-existing hypothesis – that the admixture between Neandertals and modern humans (of which UI is the best and chronologically earliest example) happened in the Middle East. The origin of this belief is an observation that Neandertal ancestry is found in all of modern non-Africans. Under an out-of-Africa scenario, this means that the admixture event occurred prior to the split into West and East Eurasians, or coastal and inland groups, somewhere in the Middle East, the closest to Africa geographic location in which Neandertal populations are attested archaeologically. But then how come that the putative “Basal Eurasian” ancestry component that modern Europeans presumably obtained in the past 10,000 years from the Middle East is not found in UI and its Eurasian relatives? An awkward answer offered by Fu et al. 2014 is that it had split from the founding East Eurasian population in the Middle East, prior to the split of UI from the latter, stayed in the Middle East and did not expand into broader West Eurasia until the Neolithic.
Out-of-America offers a much more elegant solution to this conundrum. It’s well-known that Amerindians are unique among “East Eurasians” in having “West Eurasian” ancestry. This is especially puzzling as Amerindians are geographically more removed and isolated from Europeans and demographically more depressed than, for instance, East Asians. Amerindians are consistently closer to modern Europeans than East Asians, Onge or Papuans. They are also consistently closer to ancient West Eurasians such as Mal’ta (MA-1), La Brana or any other ancient European hunter-gatherer population than modern East Asians, Papuans or Onge. However, ancient East Eurasians such as UI, which predates MA-1 by 20,000 years, are a) as close to MA-1 as they are to modern East Eurasians and b) closer to them than to modern West Eurasians. This means that Amerindians are specially related to West Eurasians through the latter’s “Basal Eurasian” ancestry. Unlike any of the Old World populations, they “play” in both major non-African clusters – “Basal Eurasian” and “Ancient North Eurasian” (ANE). This also means that “Basal Eurasian” ancestry is the “same” as “Ancient North Eurasian” (ANE) ancestry as two geographically distinct Old World offshoots of a underlying Amerindian population. Furthermore, this means that Amerindians branched off from other non-Africans before UI branched off from Eurasians. This is why, although UI and modern Amerindians are separated by a whopping 45,000 year time period, genetically modern Amerindians are only some 300 generations (roughly 9000 years if we follow Fu et al. (2014) in taking a human generation to be 29 years long) further away from Neandertals than UI.
This out-of-America scenario fits a secondary, evidence-based hypothesis voiced but dismissed by Raghavan et al. (2013):
“…If the gene flow direction was from Native Americans into western Eurasians it would have had to spread subsequently to European, Middle Eastern, south Asian and central Asian populations, including MA-1 before 24,000 years ago.”
In the light of the UI data, this out-of-America migration must have occurred prior to 45,000 years. Raghavan et al.’s (2013) main hypothesis that gene flow went from MA-1-like population into Amerindians, rather than the other way around, is also predicated on the assumption that West Eurasians is an “unadmixed” population. Lazaridis et al. 2014 show that Europeans are very much admixed. The UI study clearly shows that the special West Eurasian affinity of Amerindians could not have resulted from gene flow from West Eurasia in the aftermath of the separation of proto-Amerindians from East Asians as what makes West Eurasians distinct from East Eurasians is not the Ancient North Eurasian (ANE) component that they share with Amerindians but the “Basal Eurasian” component. The latter predates the UI times but it’s expanded from the Middle East into greater West Eurasia only in the Neolithic, which means this happened by the time America had already been peopled. Similarly, the “Basal Eurasian” component could not have been brought to Europe from America in Neolithic times, as MA-1 at 24,000 YBP already draws closer to both Amerindians and to West Eurasians. The reason MA-1 and La Brana are as close to UI as modern East Eurasians, while modern West Eurasians are clearly different from modern East Eurasians is due to the founding Amerindian migration followed by a split into two geographically distinct ancient populations – “Ancient North Eurasian” (ANE) and “Basal Eurasian”, or, better to say, “Ancient East Eurasian” (MA-1, UI, La Brana, Amerindians, East Asians, Papuans, etc.) and “Ancient West Eurasian” (modern West Eurasians and Amerindians). The hypothesis whereby West Eurasians are a product of mixture between two locally differentiated Amerindian-like components that entered Europe first at about 40,000 years from the northeast (foraging ANEs), then at about 10,000 years from the southeast (farming “Basal Eurasians”) and then, again, at 5,000 YBP (ANE again, now brought by Indo-European pastoralists), while East Eurasians (attested in UI) descend from only one Amerindian-like population fits the data. (This interpretation is only partially consistent with the three-way model of European genetic origins in Lazaridis et al. 2014.)
One of the reasons that Raghavan et al. (2013) dismissed the Amerindian source of admixture in West Eurasians was the observation that, while Amerindians are closer to East Asians than to Papuans, Mal’ta (MA-1) at 24,000 YBP is not closer to East Asians than to Papuans. Fu et al. (2014) confirm that UI at 45,000 YBP is equally removed from all of East Eurasians (presumably, including Papuans). They also show that Amerindians are followed by East Asians in the degree of retention of Neandertal ancestry chunks but leave the Papuan data out. PCA establishes (see below) the greater proximity between UI, on the one hand, and Central/South Asians followed by Amerindians, with Papuans being far removed (presumably due to additional Denisovan ancestry in Papuans).
UI’s haploid profile raises additional questions. Its mtDNA is claimed to belong to mhg R. Fu et al. (2014, SI, 47) describe their finding in the following way:
“The Ust’-Ishim individual carries the following substitutions that define the R haplogroup (compared to rCRS sequence) : 73G, 263G, 750G, 1438G, 2706G, 3107d, 4769G, 7028T, 8860G, 11719A, 14766T, 15326G as well as one specific substitution not seen in any present- day population (rCRS pos: 16150).”
A quick glance at the most recent PhyloTree mtDNA build shows that none of those mutations define hg R in relationship to N, M or L macrohaplogroups. E.g., mhg R is different from mhg N by T12705C and T16223C but those sites are not listed by Fu et al. as R-defining. The authors published mutations that separate the UI sequence from the Cambridge Reference Sequence, which is the H subset the R clade. (A special “thank you” goes to Gisele Horvat for having a thoughtful e-mail discussion of this matter with me, as well as to an anonymous commenter below.)
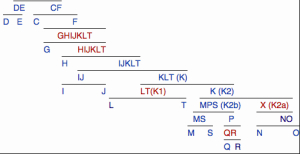
Although Fu et al. strangely defined the UI sequence against its own subclade, let’s assume that it’s indeed belongs to mhg R. While UI is the oldest known modern human sample, its mtDNA falls within the youngest, as defined by existing mtDNA phylogenies, macrohaplogroup. Notably, all of most ancient DNA samples from a wide range of geographic locations belong to the same mhg R: Kostenki in the Russian steppe at 32,000 BP is hg U, Tianyuan in south China at 40,000 is hg B, Mal’ta in South Siberia at 24,000 YBP is hg U. Presumably older mhgs N and M haven’t been found in the earliest samples. Mhg R enjoys the widest worldwide distribution now as it apparently did 45-40,000 years ago. Although autosomally UI, as we have seen, is closer to East Eurasians than to West Eurasians, mtDNA hg R is prominent in Europe (the oldest European mtDNA haplogroup is U, which is part of mhg R), while mhgs M and N are barely found in Europe and have mostly East Eurasian distribution. One intriguing exception is hg X that is somewhat frequent in the Middle East and North Africa and then in North America. In Europe it shows up only from Neolithic times and is pervasive but thinly spread across the continent. It’s distribution and time of origin in Europe fits well with the distribution of the “Basal Eurasian” autosomal component, and, as I argued above, Amerindians seem to partake in the “Basal Eurasian” component as well, just like they have mtDNA hg X. UI’s Y-DNA belongs to hg K (xLT), according to Fu et al. According to Gregory Magoon, it shares several mutations with the newly discovered, among Dravidian-speaking Telugu in South Asia, Y-DNA hg X (also known as K2a or K-M2335), which supposedly leads to the main East Asian NO cluster of lineages. The NO cluster is not found in the Americas. The main Amerindian hgs P and Q, which are rare in Asia, the most dominant European hg R (attested in MA-1 but reintroduced to Europe en masse by Indo-Europeans) as well as Papua New Guinean/Australian hgs M and S are currently thought of as belonging to the MPS (also known as K2b) cluster (see below).
This means that UI’s Y-DNA is not on the same phylogenetic path as the most frequent Amerindian and West Eurasian Y-DNA lineages but does represent an “ancestor” for the East Asian cluster. At the same time, hg K is downstream from hg C, which is the second Amerindian haplogroup attested in East Eurasia and the Sahul and, as C6, in ancient remains such as La Brana in Europe. Since the New World populations have both one of the most upstream non-African Y-DNA haplogroup, hg C, and one of the most downstream ones, hgs P and Q, but nothing in-between (no DE, no GHIJKLT, no IJKLT, no KLT and no K), it’s likely that hgs C and P are connected through a different phylogenetic path than hgs C and X (xNO).
Both Y-DNA and mtDNA are therefore consistent with the pattern seen in the autosomes but the volatility of haploid frequencies at times obfuscates ancient population relationships.




Hello
I am always impressed by your ability to interpret absolutely anything to fit your theory. 🙂
The mutations are defining haplogroup R with respect to the Cambridge Reference Sequence, which belongs to a H, itself a branch of R. Hence the substitutions listed are *ancestral* to the reference sequence mt hg. Defining mutations for R under N are not mentioned because the reference sequence and Ust’-Ishim do not differ with respect to them, both being R.
Ok, although your profile does not fit my “ideal contributor” profile, I’ll let this one slide because you wrote something meaningful. You are right! After 40 years of mtDNA research, they managed to define mhg R against its own subclade, hg H. It didn’t even occur to me. Totally Onion-worthy! 🙂
German, are you familiar with the Spirit Cave mummies from Nevada analyzed by Erv Taylor in 1996 which were dated at 11.5 kya and had European DNA? Or the Lovelock Cave mummies from Nevada whom the Paiute called the Si-Te-Cah whom were an 8 foot tall red-haired tribe in the Paiute oral history and the Paiute genocided them? Or the Florida bog mummies from Titusville Florida dated at 9500 bp buried in a similar manner as Danish burials and their DNA was the same as the Spirit Cave Nevada mummies, or Haplogroup X? Other Florida underwater burials were dated at 14000 bp and were of the same peoples.
Have you heard of the Mandan tribe of Amerindians with their blonde and red hair and blue and grey eyes? Or the Catalina Island California graveyard with anomalous skeletons of 3,781 people with different features including blonde hair and some were around 9 feet tall?
See: ‘The Ancient Giants Who Ruled America’ by Richard J. Dewhurst
‘The Lost Colonies of Ancient America’ by Frank Joseph
Do you have any thoughts on this and how it relates to the origins?
Have you read a book titled: ‘Lemurian Scrolls’, and if so, what are your thoughts on that origins story? It is less boring than the two standard narratives in this culture.