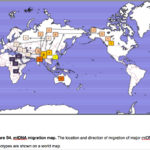
World Science en Route from Out-of-Africa to Out-of-America: First Stop is Out-of-Asia
bioRxiv doi: https://doi.org/10.1101/101410 Yuan, Dejian, Xiaoyun Lei, Yuanyuan Gui, Zuobin Zhu, Dapeng Wang, Jun Yu, and Shi Huang Modern Human Origins: Multiregional Evolution of Autosomes and East Asia Origin of Y and mtDNA Recent studies have established that genetic diversities are mostly maintained…

Peruvian Amerindians Have Strongest Genetic Ties to Archaic Hominins
Molecular Biology and Evolution (advance publication, October 18, 2016) Signatures of archaic adaptive introgression in present-day human populations Racimo, Fernando, Davide Marnettob, and Emilia Huerta-Sánchez Comparisons of DNA from archaic and modern humans show that these groups interbred, and in…
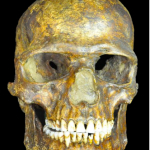
Ancient Kostenki 14 (Markina Gora) DNA: A Glimpse into a Population on Its Way from America to Africa
Science DOI: 10.1126/science.aaa0114 Genomic Structure in Europeans Dating Back at Least 36,200 Years Andaine Seguin-Orlando, Thorfinn S. Korneliussen, Martin Sikora, Anna-Sapfo Malaspinas, Andrea Manica, Ida Moltke, Anders Albrechtsen, Amy Ko, Ashot Margaryan, Vyacheslav Moiseyev, Ted Goebel, Michael Westaway, David Lambert,…
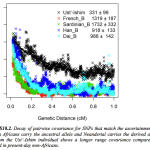
Ancient Ust’-Ishim DNA as Seen From the Americas
Nature 514, 445–449 (23 October 2014) doi:10.1038/nature13810 Genome sequence of a 45,000-year-old modern human from western Siberia Qiaomei Fu, Heng Li, Priya Moorjani, Flora Jay, Sergey M. Slepchenko, Aleksei A. Bondarev, Philip L. F. Johnson, Ayinuer Aximu-Petri, Kay Prufer, Cesare de Filippo,…
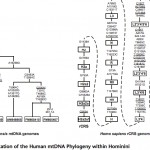
A Closer Look at Human and Neandertal Mitogenomes
In one of my comments on Anthrogenica.com, I mistakenly denied that Behar et al. (2012) reported the two mutations A2758G and G7146A shared between late European Neandertals and the modern human L2’3’4’5’6 clade. As a reminder, A2758G and G7146A were highlighted in…
The End of Out-of-Africa: A Copernican Reassessment of the Patterns of Genetic Variation in the Old World
Over at Anthrogenica, I’ve been having some heated (as always) but this time also productive discussions regarding the interpretation of currently available genetic evidence. In the following I will sketch out a hypothesis that increasingly makes sense to me. 1….
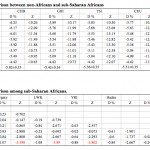
Neandertal Admixture in Africans: A Back-Migration to Sub-Saharan Africa Confirmed
Genome Biology and Evolution 2013 Oct 25. Apparent Variation in Neanderthal Admixture among African Populations is Consistent with Gene Flow from non-African Populations. Shuoguo Wang, Joseph Lachance, Sarah Tishkoff, Jody Hey, and Jinchuan Xing Abstract Recent studies have found evidence of…
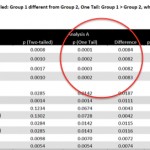
Archaic Admixture in Africa, a Final Solution for Cultural Anthropology and the New World Roots of the Oldest Dog: News from Around the Web
I haven’t been blogging for a while because of a new demanding leadership job. It will keep me busy, so I’m shifting to more of a bite-size blog post format. In the meantime, I’ve been active on Gisele Horvat’s Human…
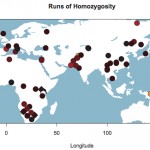
Clicks and Genes: Linguistic and Genetic Perspectives on Khoisan Prehistory
Science DOI: 10.1126/science.1227721 Genomic Variation in Seven Khoe-San Groups Reveals Adaptation and Complex African History Carina M. Schlebusch, Pontus Skoglund, Per Sjödin, Lucie M. Gattepaille, Dena Hernandez, Flora Jay, Sen Li, Michael De Jongh, Andrew Singleton, Michael G. B. Blum,…

Rare Haplotypes in Koreans: Evidence for a Back Migration into Africa?
CAMDA 2012 Conference, July 13, 2012 Rare Haplotypes in the Korean Population Sepp Hochreiter, Gunter Klambauer, Gundula Povysil, and Djork‐Arné Clevert Link (Full Text PDF) These days you never know where research related to modern human origins will be reported…
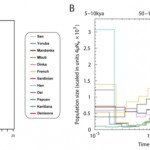
A High Coverage of the Denisovan Hominin
Science 30 August 2012 DOI: 10.1126/science.1224344 A High-Coverage Genome Sequence from an Archaic Denisovan Individual Matthias Meyer, Martin Kircher, Marie-Theres Gansauge, Heng Li, Fernando Racimo, Swapan Mallick, Joshua G. Schraiber, Flora Jay, Kay Prüfer, Cesare de Filippo, Peter H. Sudmant,…

Archaic Introgression and the Derived Nature of African Lineages at STAT2 Gene
American Journal of Human Genetics 91, 2012, 265-274. http://dx.doi.org/10.1016/j.ajhg.2012.06.015. A Haplotype at STAT2 Introgressed from Neanderthals and Serves as a Candidate of Positive Selection in Papua New Guinea Fernando L. Mendez, Joseph C. Watkins, and Michael F. Hammer. Signals of…
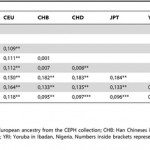
The Xavante Indians and Genetic Divergence Between American Indians and Africans
PLoS ONE 7 (8): e42702. doi:10.1371/journal.pone.0042702 Genome-Wide Analysis in Brazilian Xavante Indians Reveals Low Degree of Admixture Patricia C. Kuhn, Andréa R. V. Russo. Horimoto, José Maurício Sanches, João Paulo B. Vieira Filho, Luciana Franco, Amaury Dal Fabbro, Laercio Joel…
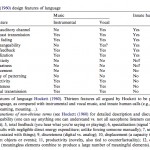
The Evolution of Language and Music
Cognition 100 (2006): 173-215 http://dx.doi.org/10.1016/j.cognition.2005.11.009 The Biology and Evolution of Music: A Comparative Perspective Fitch, W. Tecumseh. Studies of the biology of music (as of language) are highly interdisciplinary and demand the integration of diverse strands of evidence. In this…

Social Anthropology and the Bantu Expansion
Razib is now officially a fantasy science blogger. When he recently called his readers “stupid, ignorant or lazy” and put up a stringent comments policy (I bet inspired by my own) rallying them to show their “A-game,” I knew it…
A Seismic Shift in Human Origins Research, or a Downward Slide?
A new research paper is out which has created a lot of media buzz. “Evolutionary History and Adaptation from High-Coverage Whole-Genome Sequences of Diverse African Hunter-Gatherers, “ by Joseph Lachance et al. reports “archaic admixture” in three African hunter-gathering populations…

Khoisans Are Genetically Admixed and Not Basal to Other Humans, Hadza Are Recently Admixed
In my last post I discussed the ways in which phylogenetic trees constructed on the basis of genetic data confound population relationships derived from common descent and from admixture. Unless admixture is taken into account, the sampled populations may end…
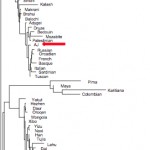
How to Interpret Patterns of Genetic Variation? Admixture, Divergence, Inbreeding, Cousin Marriage
Two different but important population genetics papers have come out. One is Steven Bray et al. (2010) “Signatures of Founder Effects, Admixture, and Selection in the Ashkenazi Jewish Population.” The other one is Isabel Alves et al. (2012) “Genomic Data…
Out-of-Africa in the Mid-Pleistocene: A New Interdisciplinary Paradigm or a New Myth?
In the comments section on this blog, Dienekes raises the issue of interdisciplinary support for the out-of-America theory. Since I’m a big proponent of interdisciplinarity, the seeming convergence of genetics, archeology and paleobiology on the origin of modern humans in…
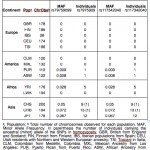
Neandertal Admixture in microRNA Genes
Molecular Biology and Evolution (27 January 2012), doi:10.1093/molbev. An Ancestral miR-1304 Allele Present in Neanderthals Regulates Genes Involved in Enamel Formation and Could Explain Dental Differences with Modern Humans Lopez-Valenzuela, Maria, Oscar Ramirez, Antonio Rosas, Samuel Garcıa-Vargas, Marco de la…

Howler Monkeys, Neandertals, Pygmies, Khoisans and More: Society for Molecular Biology and Evolution 2012
As I write, Society for Molecular Biology and Evolution (SMBE) is conducting its annual meetings in Dublin, Ireland. Dienekes has many useful pullouts from the available abstracts. I will make short comments on a few of them, plus bring in…
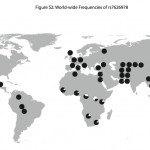
The Pygmy Enigma: Biology, Population Genetics and Linguistics
PLoS Genetics 8 (4): e1002641. doi:10.1371/journal.pgen.1002641 Patterns of Ancestry, Signatures of Natural Selection, and Genetic Association with Stature in Western African Pygmies Jarvis, Joseph P., Laura B. Scheinfeldt, Sameer Soi, Charla Lambert, Larsson Omberg, Bart Ferwerda, Alain Froment, Jean-Marie Bodo,…
Phonemic Diversity and out-of-Africa Again: The Myth is Gaining a Momentum
PLoS ONE 7(4): e35289. doi:10.1371/journal.pone.0035289 Dating the Origin of Language Using Phonemic Diversity Perreault, Charles, and Sarah Mathew. Abstract Language is a key adaptation of our species, yet we do not know when it evolved. Here, we use data on…
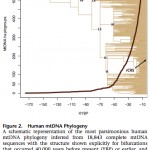
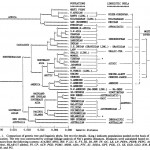
Between Behar et al. 2012 and Johnson et al. 1983: The Mitochondrial DNA Tree Comes of Age but Remains a Blunt Tool for Human Evolutionary History
American Journal of Human Genetics, Volume 90, Issue 4, 675-684, 6 April 2012 doi:10.1016/j.ajhg.2012.03.002 A “Copernican” Reassessment of the Human Mitochondrial DNA Tree from its Root Behar, Doron M., Mannis van Oven, Saharon Rosset, Mait Metspalu, Eva-Liis Loogvali, Nuno M….

The Origin of mtDNA haplogroup B: 9-bp deletion in America, Asia and Africa
PLoS ONE 7(2) 2012: e32179. doi:10.1371/journal.pone.0032179 Complete Mitochondrial DNA Analysis of Eastern Eurasian Haplogroups Rarely Found in Populations of Northern Asia and Eastern Europe Derenko M., Malyarchuk B., Denisova G., Perkova M., Rogalla U., et al. Abstract. With the aim…
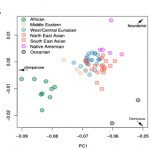
American Indians, Neanderthals and Denisovans: Insights from PCA Views
Dienekes posted a SNP PCA showing the relative position of a sample of modern human populations from the Harvard HGDP along the axes formed by Chimpanzees, Denisovans and Neanderthals. On the broad-view PCA, the red dot indicates Chimpanzees, the green…


Recent Comments